Targeting the temporal dynamics of hypoxia-induced tumor-secreted factors halts tumor migration
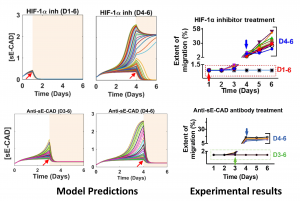
Targeting microenvironmental factors that foster migratory cell phenotypes is a promising strategy for halting tumor migration. However, lack of mechanistic understanding of the emergence of migratory phenotypes impedes pharmaceutical drug development. Using our 3D microtumor model with tight control over tumor size, we recapitulated the tumor size-induced hypoxic microenvironment and emergence of migratory phenotypes in microtumors from epithelial breast cells and patient-derived primary metastatic breast cancer cells, mesothelioma cells, and lung cancer xenograft cells (PDX). The microtumor models from various patient-derived tumor cells and PDX cells revealed upregulation of tumor-secreted factors including matrix metalloproteinase-9 (MMP9), fibronectin (FN), and soluble E-cadherin (sE-CAD), consistent with clinically reported elevated levels of FN and MMP9 in patient breast tumors compared to healthy mammary glands. Secreted factors in the conditioned media of large microtumors induced a migratory phenotype in non-hypoxic, non-migratory small microtumors. Subsequent mathematical analyses identified a two-stage microtumor progression and migration mechanism whereby hypoxia induces a migratory phenotype in the initialization stage which then becomes self-sustained through a positive feedback loop established among the tumor-secreted factors. Computational and experimental studies showed that inhibition of tumor-secreted factors effectively halts microtumor migration despite tumor-to-tumor variation in migration kinetics, while inhibition of hypoxia is effective only within a time window and is compromised by tumor-to-tumor variation, supporting our notion that hypoxia initiates migratory phenotypes but does not sustain it. In summary, we show that targeting temporal dynamics of evolving microenvironments, especially tumor-secreted factors during tumor progression, can halt tumor migration.
Singh M, Tian XJ, Donnenberg VS, Watson AM, Zhang JY, Stabile LP, Watkins SC, Xing J, Sant S. (2019) Targeting the temporal dynamics of hypoxia-induced tumor-secreted factors halts tumor migration. Cancer Res. DOI: 10.1158/0008-5472.CAN-18-3151
 |
 |
| Shilpa Sant, PhD | Jianhua Xing, PhD |





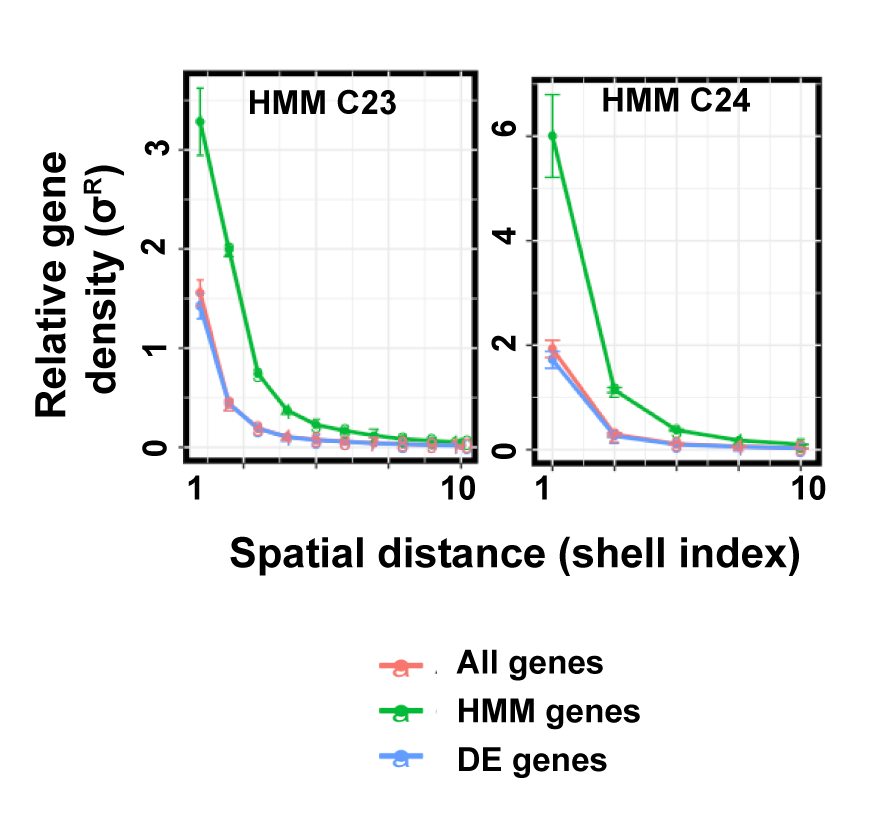 Cellular responses to environmental stimulation are often accompanied by changes in gene expression patterns. Genes are linearly arranged along chromosomal DNA, which folds into a three-dimensional structure. The chromosome structure affects gene expression activities and is regulated by multiple events such as histone modifications and DNA binding of transcription factors. A basic question is how these mechanisms work together to regulate gene expression. In this study, we analyzed temporal gene expression patterns in the context of chromosome structure both in a human cell line under TGF-β treatment and during mouse nervous system development. In both cases, we observed that genes regulated by common transcription factors have an enhanced tendency to be spatially close. Our analysis suggests that spatial co-localization of genes may facilitate the concerted gene expression.
Cellular responses to environmental stimulation are often accompanied by changes in gene expression patterns. Genes are linearly arranged along chromosomal DNA, which folds into a three-dimensional structure. The chromosome structure affects gene expression activities and is regulated by multiple events such as histone modifications and DNA binding of transcription factors. A basic question is how these mechanisms work together to regulate gene expression. In this study, we analyzed temporal gene expression patterns in the context of chromosome structure both in a human cell line under TGF-β treatment and during mouse nervous system development. In both cases, we observed that genes regulated by common transcription factors have an enhanced tendency to be spatially close. Our analysis suggests that spatial co-localization of genes may facilitate the concerted gene expression.
