Spatial clustering and common regulatory elements correlate with coordinated gene expression
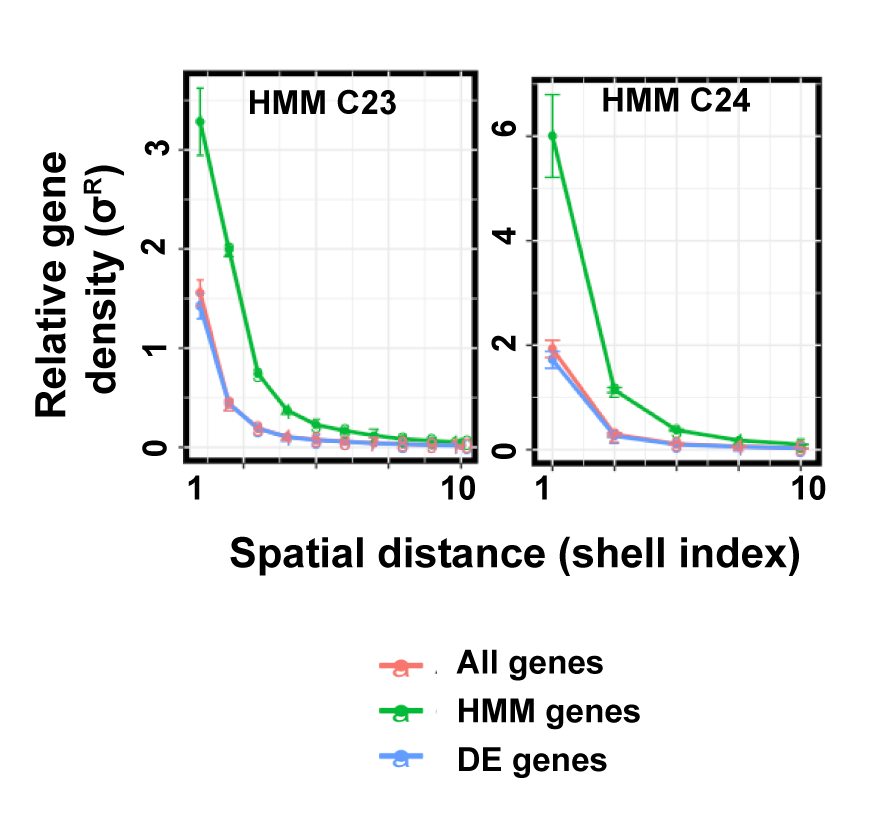 Cellular responses to environmental stimulation are often accompanied by changes in gene expression patterns. Genes are linearly arranged along chromosomal DNA, which folds into a three-dimensional structure. The chromosome structure affects gene expression activities and is regulated by multiple events such as histone modifications and DNA binding of transcription factors. A basic question is how these mechanisms work together to regulate gene expression. In this study, we analyzed temporal gene expression patterns in the context of chromosome structure both in a human cell line under TGF-β treatment and during mouse nervous system development. In both cases, we observed that genes regulated by common transcription factors have an enhanced tendency to be spatially close. Our analysis suggests that spatial co-localization of genes may facilitate the concerted gene expression.
Cellular responses to environmental stimulation are often accompanied by changes in gene expression patterns. Genes are linearly arranged along chromosomal DNA, which folds into a three-dimensional structure. The chromosome structure affects gene expression activities and is regulated by multiple events such as histone modifications and DNA binding of transcription factors. A basic question is how these mechanisms work together to regulate gene expression. In this study, we analyzed temporal gene expression patterns in the context of chromosome structure both in a human cell line under TGF-β treatment and during mouse nervous system development. In both cases, we observed that genes regulated by common transcription factors have an enhanced tendency to be spatially close. Our analysis suggests that spatial co-localization of genes may facilitate the concerted gene expression.
Zhang J, Chen H, Li R, Taft DA, Yao G, Bai F, Xing J. (2019) Spatial clustering and common regulatory elements correlate with coordinated gene expression. PLoS Computational Biology 15(3):e1006786
