Mr. Nuri Has (left), Dr. Ivet Bahar (middle), and Dr. Sondan Durukanoglu Feyiz (right)
The 15th Annual Kadir Has Awards were held on Friday, March 22nd in Istanbul, Turkey. Member of the Science Academy, Dr. Ivet Bahar, received the Kadir Has “Outstanding Achievement Award” for her contributions to the development of theoretical and computational models for explaining the functional dynamics of biomolecular systems as well as mentoring and teaching a new wave of scientists. She was presented with the award by the Chairman of the Board of Trustees of Kadir Has University, Mr. Nuri Has, the Kadir Has Foundation President, Mr. Can Has, and the President of Kadir Has University, Dr. Sondan Durukanoglu Feyiz.
The Kadir Has Awards seek to recognize the outstanding accomplishments that Turkish scientists have made at the national and international level and to promote people and institutions that have contributed to the development of society.
For more information, please visit http://kadirhasvakfi.org/en/.






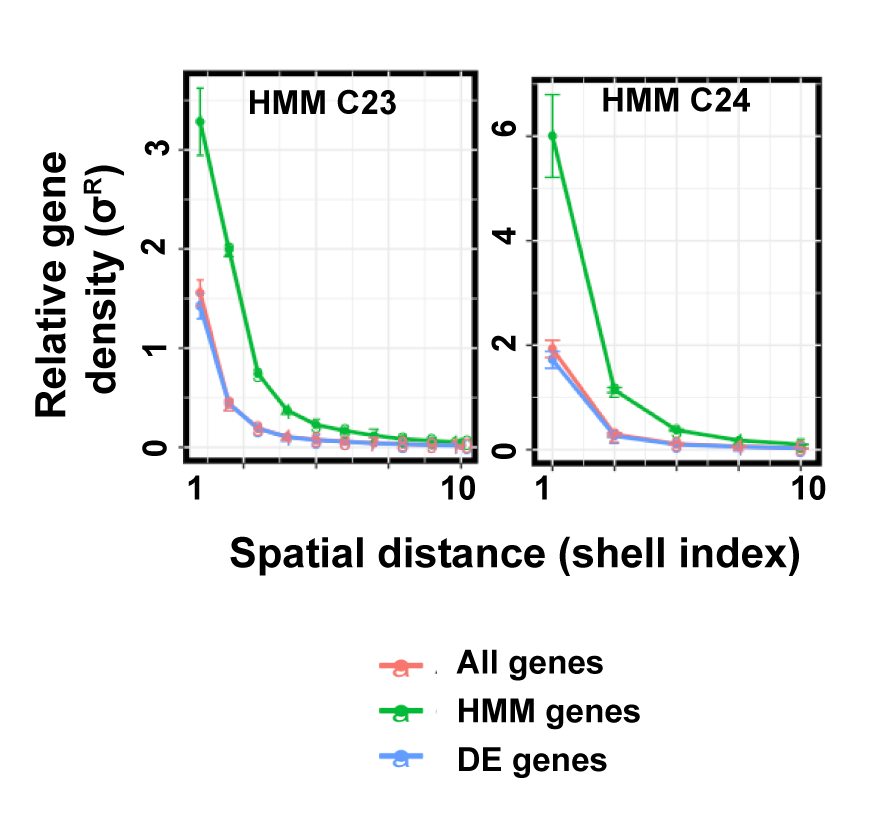 Cellular responses to environmental stimulation are often accompanied by changes in gene expression patterns. Genes are linearly arranged along chromosomal DNA, which folds into a three-dimensional structure. The chromosome structure affects gene expression activities and is regulated by multiple events such as histone modifications and DNA binding of transcription factors. A basic question is how these mechanisms work together to regulate gene expression. In this study, we analyzed temporal gene expression patterns in the context of chromosome structure both in a human cell line under TGF-β treatment and during mouse nervous system development. In both cases, we observed that genes regulated by common transcription factors have an enhanced tendency to be spatially close. Our analysis suggests that spatial co-localization of genes may facilitate the concerted gene expression.
Cellular responses to environmental stimulation are often accompanied by changes in gene expression patterns. Genes are linearly arranged along chromosomal DNA, which folds into a three-dimensional structure. The chromosome structure affects gene expression activities and is regulated by multiple events such as histone modifications and DNA binding of transcription factors. A basic question is how these mechanisms work together to regulate gene expression. In this study, we analyzed temporal gene expression patterns in the context of chromosome structure both in a human cell line under TGF-β treatment and during mouse nervous system development. In both cases, we observed that genes regulated by common transcription factors have an enhanced tendency to be spatially close. Our analysis suggests that spatial co-localization of genes may facilitate the concerted gene expression.