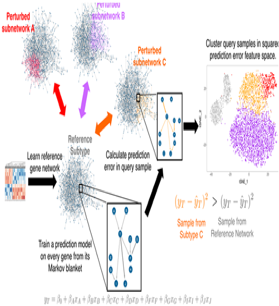
 | Buschur KL, Chikina M, Benos PV. Causal network perturbations for instance-specific analysis of single cell and disease samples.Bioinformatics. 2019 Dec 24. pii: btz949. doi: 10.1093/bioinformatics/btz949. PubMed PMID: 31873725 |
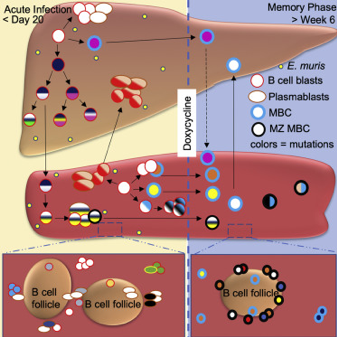 | Trivedi N, Weisel F, Smita S, Joachim S, Kader M, Radhakrishnan A, Clouser C, Rosenfeld AM, Chikina M, Vigneault F, Hershberg U, Ismail N, Shlomchik MJ. Liver Is a Generative Site for the B Cell Response to Ehrlichia muris. Immunity. 2019 Dec 17;51(6):1088-1101.e5. doi: 10.1016/j.immuni.2019.10.004. Epub 2019 Nov 12. PubMed PMID: 31732168 |
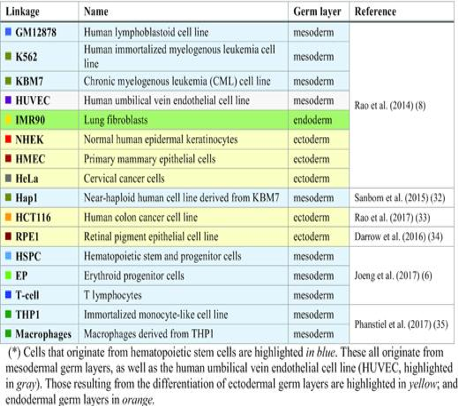 | Zhang S. Chen F, Bahar I. Differences in the intrinsic spatial dynamics of the chromatin contribute to cell differentiation. Nucleic Acids Res. 2019 Dec 12. pii: gkz1102. doi: 10.1093/nar/gkz1102. PubMed PMID:31828312 |
 | Belovich AN, Aguilar JI, Mabry SJ, Cheng MH, Zanella D, Hamilton PJ, Stanislowski DJ, Shekar A, Foster JD, Bahar I, Matthies HJG, Galli A. A network of phosphatidylinositol (4,5)-bisphosphate (PIP2) binding sites on the dopamine transporter regulates amphetamine behavior in Drosophila Melanogaster. Mol Psychiatry. 2019 Dec 3 doi: 10.1038/s41380-019-0620-0. Pub Med PMID: 31796894 |
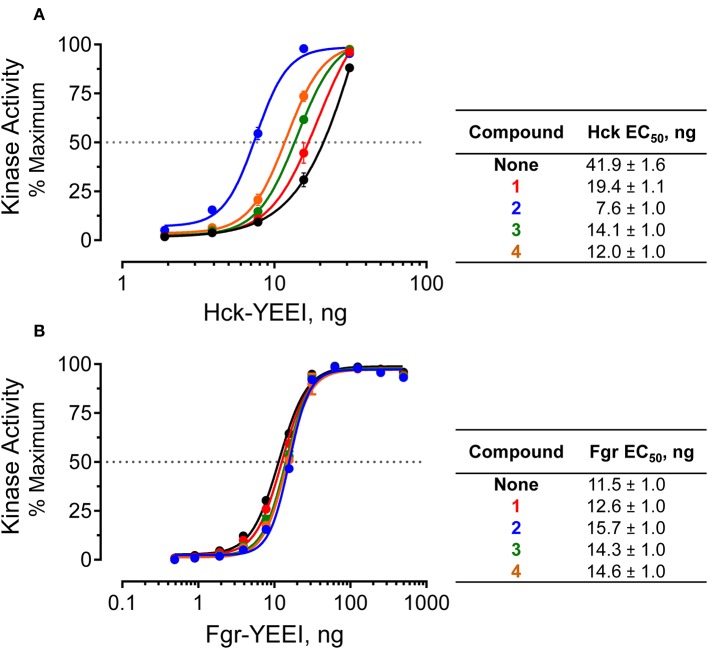 | Dorman HR, Close D, Wingert BM, Camacho CJ, Johnston PA, Smithgall TE. Discovery of Non-peptide Small Molecule Allosteric Modulators of the Src-family Kinase, Hck.Front Chem. 2019 Nov 28 doi: 10.3389/fchem.2019.00822. eCollection 2019. PubMed PMID: 31850311 |
| Zhang Y, Doruker P, Kaynak B, Zhang S, Krieger J, Li H, Bahar I. Intrinsic dynamics is evolutionarily optimized to enable allosteric behavior. Curr Opin Struct Biol. 2019 Nov 27 doi: 10.1016/j.sbi.2019.11.002. PubMed PMID: 31785465 | |
 | Lowe KE, Regan EA, Anzueto A, Austin E, Austin JHM, Beaty TH, Benos PV,..., Silverman EK and Crapo JD. (2019) COPDGene ® 2019: Redefining the Diagnosis of Chronic Obstructive Pulmonary Disease. Chronic Obstr Pulm Dis. 6 (5): 384-399. PMID: 31710793. |
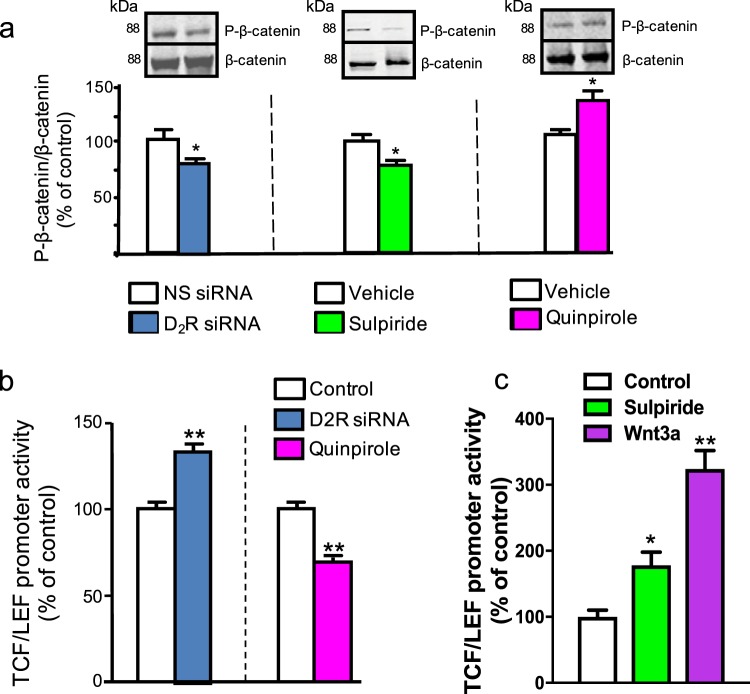 | Han F, Konkalmatt P, Mokashi C, Kumar M, Zhang Y, Ko A, Farino ZJ, Asico LD, Xu G, Gildea J, Zheng X, Felder RA, Lee REC, Jose PA, Freyberg Z, Armando I. Dopamine D2 receptor modulates Wnt expression and control of cell proliferation.Sci Rep. 2019 Nov 14;9(1):16861. doi: 10.1038/s41598-019-52528-4. PubMed PMID: 31727925 |
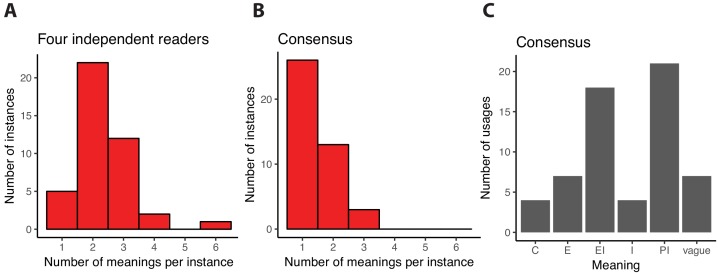 | Keeling DM, Garza P, Nartey CM, Carvunis AR. The meanings of 'function' in biology and the problematic case of de novo gene emergence. Elife. 2019 Nov 1;8. pii: e47014. doi: 10.7554/eLife.47014. PubMed PMID: 31674305 |
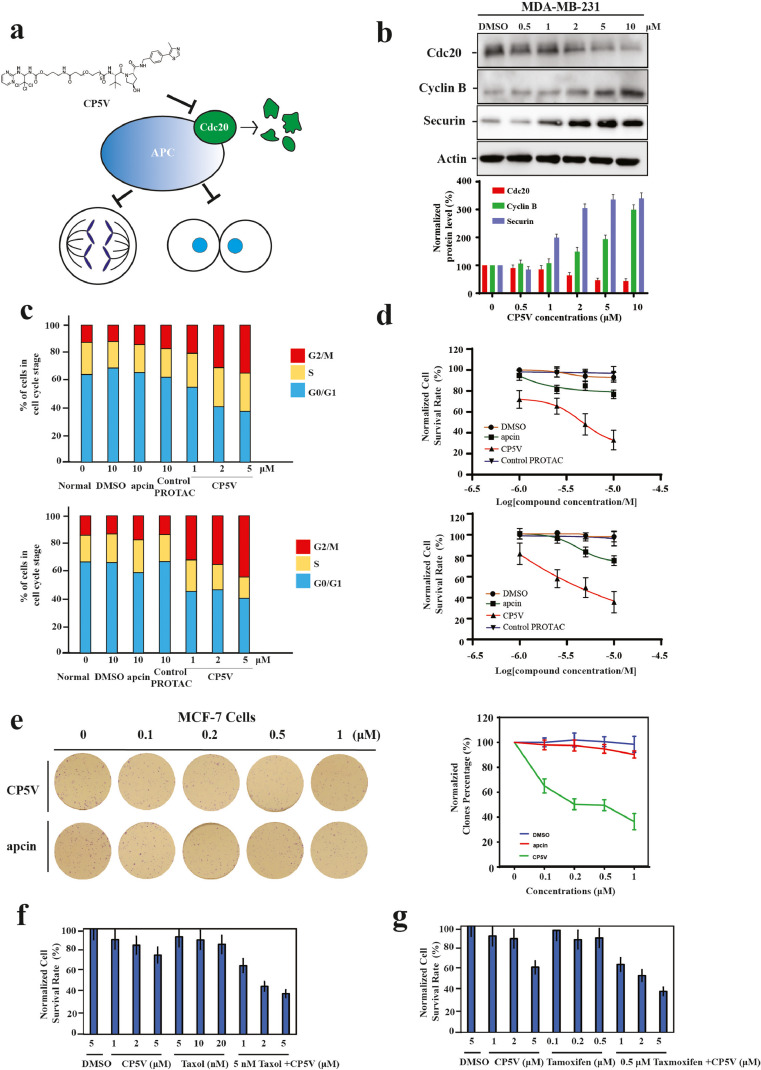 | Chi JJ, Li H, Zhou Z, Izquierdo-Ferrer J, Xue Y, Wavelet CM, Schiltz GE, Zhang B, Cristofanilli M, Lu X, Bahar I, Wan Y. A novel strategy to block mitotic progression for targeted therapy. EBioMedicine. 2019 Nov;49:40-54. doi: 10.1016/j.ebiom.2019.10.013. Epub 2019 Oct 25. PubMed PMID:31669221 |
| Berg J. Editor's note. Science. 2019 Oct 25;366(6464):432. doi: 10.1126/science.aaz9111. PubMed PMID: 31649187. | |
| Berg J. On privilege. Science. 2019 Oct 25;366(6464):401. doi: 10.1126/science.aaz8977. PubMed PMID: 31649169. | |
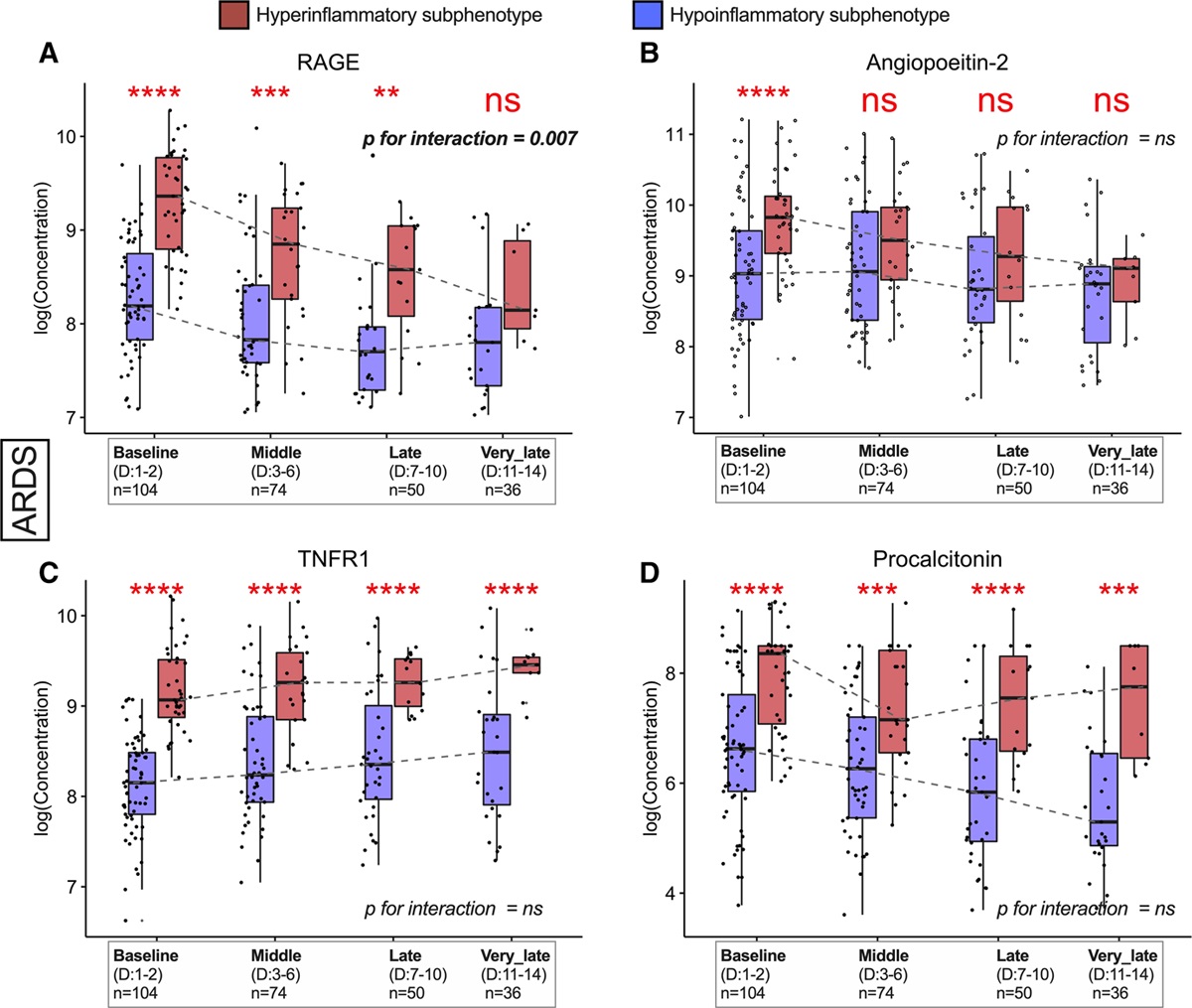 | Kitsios GD, Yang L, Manatakis DV, Nouraie M, Evankovich J, Bain W, Dunlap DG, Shah F, Barbash IJ, Rapport SF, Zhang Y, DeSensi RS, Weathington NM, Chen BB, Ray P, Mallampalli RK, Benos PV, Lee JS, Morris A, McVerry BJ. Host-Response Subphenotypes Offer Prognostic Enrichment in Patients With or at Risk for Acute Respiratory Distress Syndrome. Crit Care Med. 2019 Oct 18. doi: 10.1097/CCM.0000000000004018. [Epub ahead of print] PubMed PMID: 31634231. |
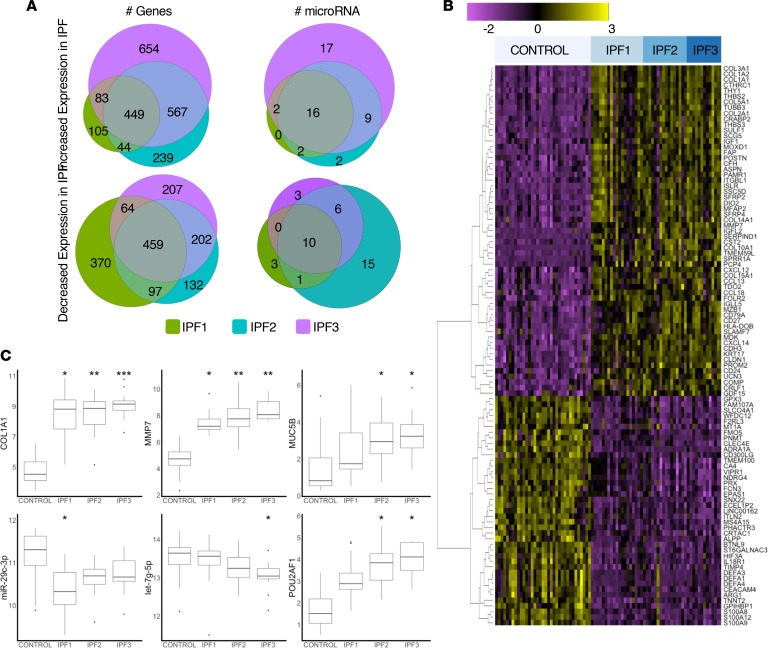 | McDonough JE, Ahangari F, Li Q, Jain S, Verleden SE, Herazo-Maya J, Vukmirovic M, Deluliis G, Tzouvelekis A, Tanabe N, Chu F, Yan X, Verschakelen J, Homer RJ, Manatakis DV, Zhang J, Ding J, Maes K, De Sadeleer L, Vos R, Neyrinck A, Benos PV, Bar-Joseph Z, Tantin D, Hogg JC, Vanaudenaerde BM, Wuyts WA, Kaminski N. (2019) Transcriptional Regulatory Model of Fibrosis Progression in the Human Lung. JCI Insight. 2019 Nov 14; 4(22):e131597. PMID: 31600171 |
 | Kohan I, Berg JM. Donald A. B. Lindberg (1933-2019). Science. 2019 Oct 4; 366(6461):37. doi: 10.1126/science.aaz3644. PubMed PMID: 31604294. |
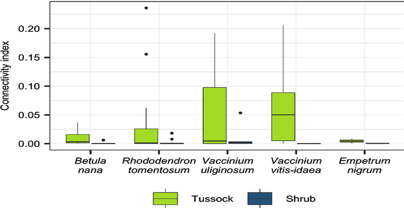 | Hewitt RE, DeVan MR, Lagutina IV, Genet H, McGuire AD, Taylor DL, Mack MC. Mycobiont contribution to tundra plant acquisition of permafrost-derived nitrogen. New Phytol. 2019 Oct 3. doi: 10.1111/nph.16235. PubMed PMID: 3150482 |
| Lee JY, Krieger JM, Li H, Bahar I. Pharmmaker: Pharmacophore modeling and hit identification based on druggability simulations. Protein Sci. 2019 Oct 1. doi: 10.1002/pro.3732. [Epub ahead of print] PubMed PMID: 31576621. | |
| Taylor DL, Espeleta HC, Kraft JD, Grant DM. Early childhood experiences and cognitive risk factors for anxiety symptoms among college students. J Am Coll Health. 2019 Sep 24:1-7. doi: 10.1080/07448481.2019.1664552. PubMed PMID:31549919 | |
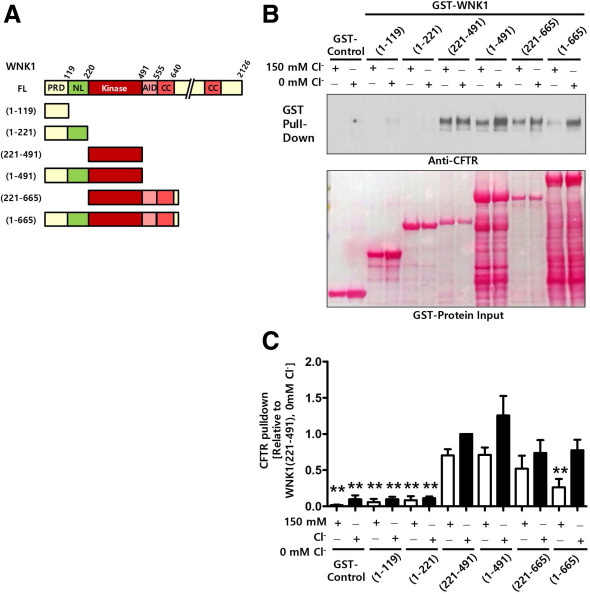 | Kim Y, Jun I, Shin DH, Yoon JG, Piao H, Jung J, Park HW, Cheng MH, Bahar I, Whitcomb DC, Lee MG. Regulation of CFTR Bicarbonate Channel Activity by WNK1: Implications for Pancreatitis and CFTR-Related Disorders. Cell Mol Gastroenterol Hepatol. 2019 Sep 24. pii: S2352-345X(19)30121-3. doi: 10.1016/j.jcmgh.2019.09.003. [Epub ahead of print] PubMed PMID: 31561038. |
| Berg J. Editorial expression of concern. Science. 2019 Sep 6;365(6457):991. doi: 10.1126/science.aaz2722. PubMed PMID: 31488679. | |
 | Berg J. Replication challenges. Science. 2019 Sep 6;365(6457):957. doi: 10.1126/science.aaz2701. PubMed PMID: 31488663. |
 | Morse C, Tabib T, Sembrat J, Buschur KL, Bittar HT, Valenzi E, JIang Y, Kass DJ, Gibson K, Chen W, Mora A, Benos PV, Rojas M, Lafyatis R. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J. 2019 Aug 22;54(2). pii: 1802441. doi: 10.1183/13993003.02441-2018. Print 2019 Aug. PubMed PMID: 31221805. |
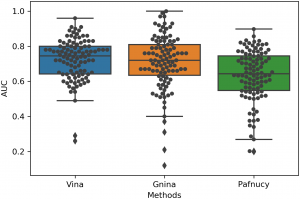 | Chen L, Cruz A, Ramsey S, Dickson CJ, Duca JS, Hornak V, Koes DR, Kurtzman T. Hidden bias in the DUD-E dataset leads to misleading performance of deep learning in structure-based virtual screening. PLoS One. 2019 Aug 20;14(8):e0220113. doi: 10.1371/journal.pone.0220113. eCollection 2019. PubMed PMID: 31430292. |
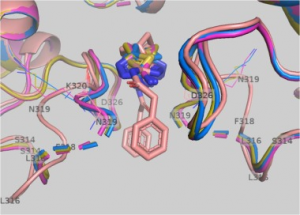 | McDermott L, Koes D, Mohammed S, Iyer P, Boby M, Balasubramanian V, Geedy M, Katt W, Cerione R. GAC inhibitors with a 4-hydroxypiperidine spacer: Requirements for potency. Bioorg Med Chem Lett. 2019 Aug 20:126632. doi: 10.1016/j.bmcl.2019.126632. [Epub ahead of print] PubMed PMID: 31474484. |
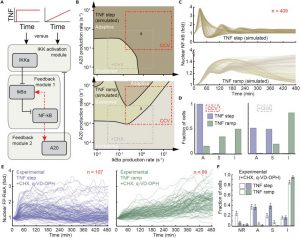 | Mokashi CS, Schipper DL, Qasaimeh MA, Lee REC. A System for Analog Control of Cell Culture Dynamics to Reveal Capabilities of Signaling Networks. iScience. 2019 Aug 8;19:586-596. doi: 10.1016/j.isci.2019.08.010. [Epub ahead of print] PubMed PMID: 31446223; PubMed Central PMCID: PMC6713801. |
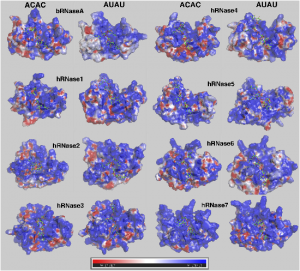 | Bafna K, Narayanan C, Chennubhotla SC, Doucet N, Agarwal PK. Nucleotide substrate binding characterization in human pancreatic-type ribonucleases. PLoS One. 2019 Aug 8;14(8):e0220037. doi: 10.1371/journal.pone.0220037. eCollection 2019. PubMed PMID: 31393891; PubMed Central PMCID: PMC6687278. |
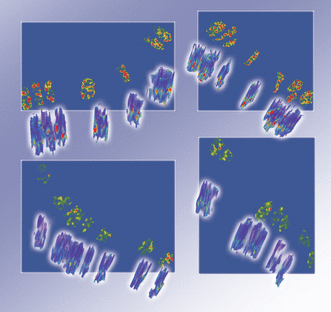 | Uttam S, Hashash JG, LaFace J, Binion D, Regueiro M, Hartman DJ, Brand RE, Liu Y. Three-Dimensional Nanoscale Nuclear Architecture Mapping of Rectal Biopsies Detects Colorectal Neoplasia in Patients with Inflammatory Bowel Disease. 2019 August. Cancer Prevention Research, 12(8):527-538 |
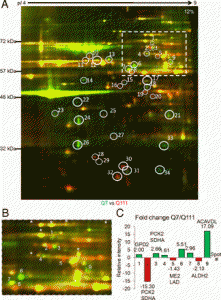 | Yablonska S, Ganesan V, Ferrando LM, Kim J, Pyzel A, Baranova OV, Khattar NK, Larkin TM, Baranov SV, Chen N, Strohlein CE, Stevens DA, Wang X, Chang YF, Schurdak ME, Carlisle DL, Minden JS, Friedlander RM. Mutant huntingtin disrupts mitochondrial proteostasis by interacting with TIM23. Proc Natl Acad Sci U S A. 2019 Aug 13;116(33):16593-16602. doi: 10.1073/pnas.1904101116. Epub 2019 Jul 25. PubMed PMID: 31346086; PubMed Central PMCID: PMC6697818. |
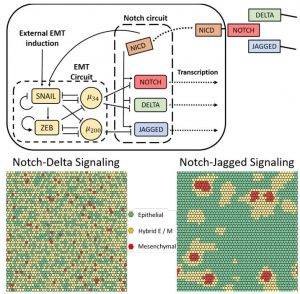 | Tripathi S, Xing J, Levine H, Jolly MK, Mathematical Modeling of Plasticity and Heterogeneity in EMT, arXiv. 25 July 2019 |
 | Liu C, Chikina M, Deshpande R, Menk AV, Wang T, Tabib T, Brunazzi EA, Vignali KM, Sun M, Stolz DB, Lafyatis RA, Chen W, Delgoffe GM, Workman CJ, Wendell SG, Vignali DAA. Treg Cells Promote the SREBP1-Dependent Metabolic Fitness of Tumor-Promoting Macrophages via Repression of CD8+ T Cell-Derived Interferon-γ. Immunity. 2019 Aug 20;51(2):381-397.e6. doi: 10.1016/j.immuni.2019.06.017. Epub 2019 Jul 23. PubMed PMID: 31350177; PubMed Central PMCID: PMC6703933. |
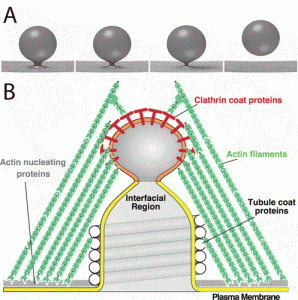 | Igoshin OA, Chen J, Xing J, Liu J, Elston TC, Grabe M, Kim KS, Nirody JA, Rangamani P, Sun SX, Wang H, Wolgemuth C. Biophysics at the coffee shop: lessons learned working with George Oster. 2019 July 22. Molecular Biology of the Cell; 30(16): 1882-1889. |
 | Berg J. A Child of Apollo. 2019 July 19. Science; 365(6450): 203. |
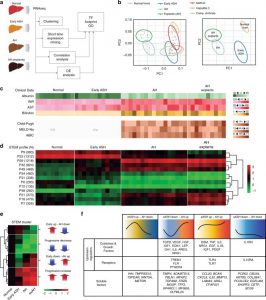 | Argemi J, Latasa MU, Atkinson SR, Blokhin IO, Massey V, Gue JP, Cabezas J, Lozano JJ, Van Booven D, Bell A, Cao S, Vernetti LA, Arab JP, Ventura- Cots M, Edmunds LR, Fondevilla C, Starkel P, Dubuquoy L, Louvet A, Odena G, Gomez JL, Aragon T, Altamirano J, Caballeria J, Jurzcak MJ, Taylor DL, Berasain C, Wahlestedt C, Monga SP, Morgan MY, Sancho-Bru P, Mathurin P, Furuya S, Lackner C, Rusyn I, Shah VH, Thursz MR, Mann J, Avila MA, Bataller R. Defective HNF4alpha-dependent gene expression as a driver of hepatocellular failure in alcoholic hepatitis. Nat Commun. 2019;10(1):3126. Published 2019 Jul 16. doi:10.1038/s41467-019-11004-3. |
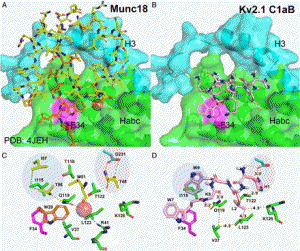 | Yeh CY, Ye Z, Moutal A, Gaur S, Henton AM, Kouvaros S, Saloman JL, Hartnett-Scott KA, Tzounopoulos T, Khannah R, Aizenman E, Camacho CJ. Defining the Kv2.1-syntaxin molecular interaction identifies a first-in-class small molecule neuroprotectant. 2019 July 15. Proceedings of the National Academy of Sciences of the United States of America. |
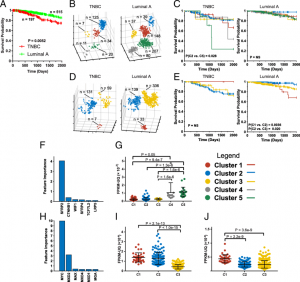 | Mandel J, Wang H, Normolle DP, Chen W, Yan Q, Lucas PC, Benos PV, Prochownik EV. Expression patterns of small numbers of transcripts from functionally- related pathways predict survival in multiple cancers. 2019 Jul 12. BMC Cancer; 19(1):686. |
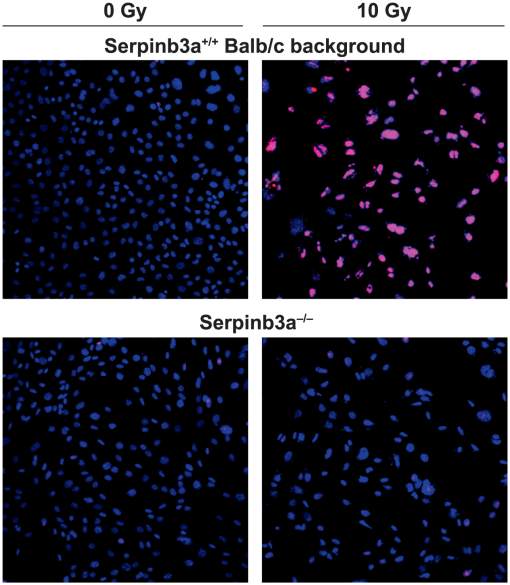 | Thermozier S, Zhang X, Hou W, Fisher R, Epperly MW, Liu B, Bahar I, Wang H, Greenberger JS. Radioresistance of Serpinb3a-/- Mice and Derived Hematopoietic and Marrow Stromal Cell Lines. Radiat Res. 2019 Sep;192(3):267-281. doi:10.1667/RR15379.1. Epub 2019 Jul 11. PubMed PMID: 31295086; PubMed Central PMCID: PMC6759811. |
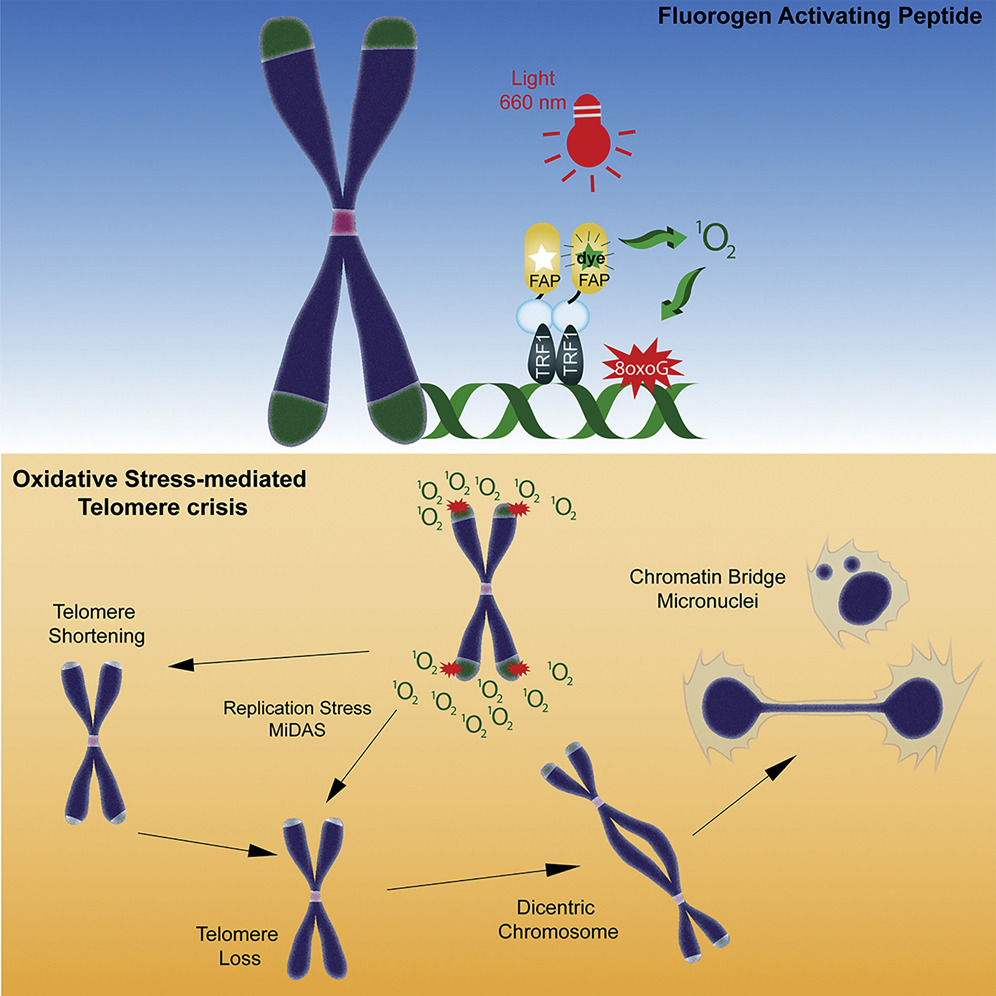 | Fouquerel E, Barnes RP, Uttam S, Watkins SC, Bruchez MP, Opresko PL. Targeted and Persistent 8-Oxoguanine Base Damage at Telomeres Promotes Telomere Loss and Crisis. 2019 July 11. Molecular Cell, 75(1): 117 - 130 |
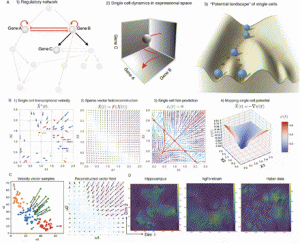 | Qiu X, Zhang Y, Yang, Hosseinzadeh S, Wang L, Yuan R, Xu S, Ma Y, Replogle J, Darmanis S, Xing J, Weissman J, Mapping Vector Field of Single Cells, BioRxiv. 2019 July 9 |
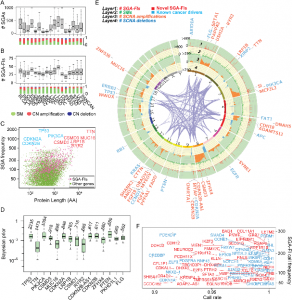 | Cai C, Cooper GF, Lu KN, Ma X, Xu S, Zhao Z, Chen X, Xue Y, Lee AV, Clark N, Chen V, Lu S, Chen L, Yu L, Hochheiser HS, Jiang X, Wang QJ, Lu X. Systematic discovery of the functional impact of somatic genome alterations in individual tumors through tumor-specific causal inference. 2019 Jul 5. PLOS Computational Biology; 15(7) |
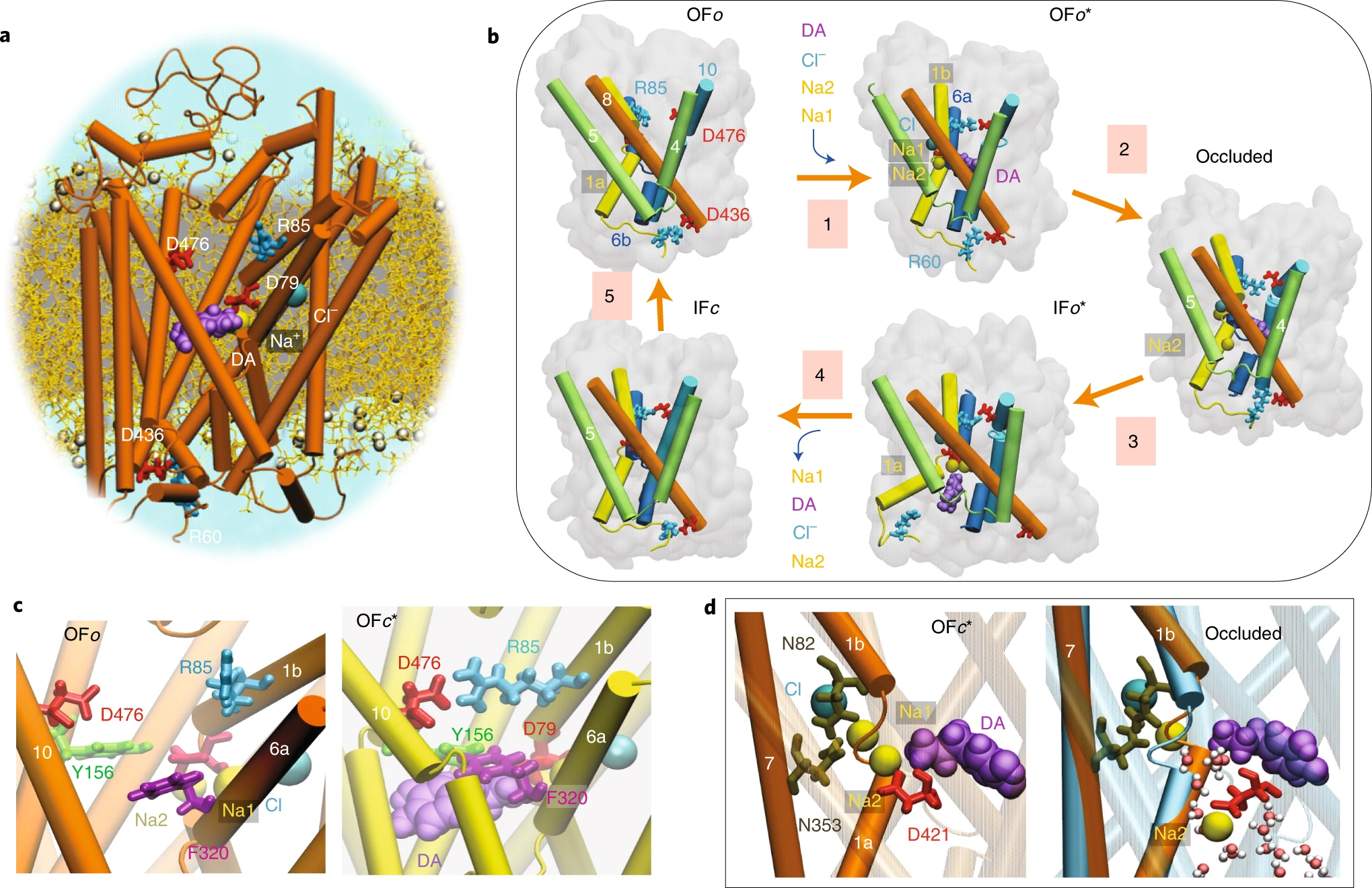 | Cheng MH, Bahar I. Monoamine transporters: structure, intrinsic dynamics and allosteric regulation. 2019 July 03. Nature Structural & Molecular Biology; 26(7): 545-556. |
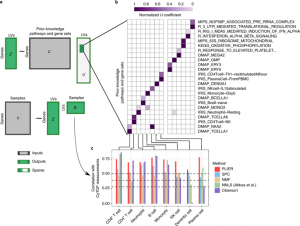 | Mao W, Zaslavsky E, Hartmann BM, Sealfon SC, Chikina M. Pathway-level information exterior (PLIER) for gene expression data. 2019 July. Nature methods; 16(7): 607-610. |
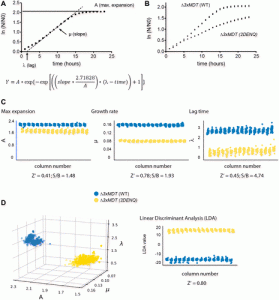 | Ngo M, Wechter N, Tsai E, Shun TY, Gough A, Schurdak ME, Schwacha A, Vogt A. A High-Throughput Assay for DNA Replication Inhibitors Based upon Multivariate Analysis of Yeast Growth Kinetics. 2019 July. SLAS Discovery; 24(6):669-681. |
| Berg J. DNA patents revisited. 2019 Jun 21. Science; 364(6446):1113. | |
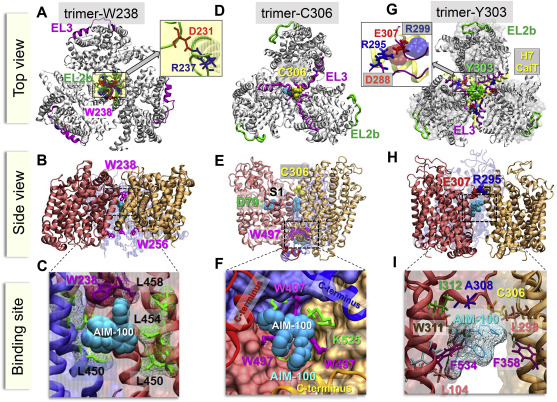 | Cheng MH, Ponzoni L, Sorkina T, Lee JY, Zhang She, SorkinA, Bahar I. Trimerization of dopamine transporter triggered by AIM-100 binding: Molecular mechanism and effect of mutations. 2019 Jun 20. Neuropharmacology; 107676. |
 | Taylor DL, Gough A, Schurdak ME, Vernetti L, Chennubhotla CS, Lefever D, Pei F, Faeder JR, Lezon TR, Stern AM, Bahar I. Harnessing Human Microphysiology Systems as Key Experimental Models for Quantitative Systems Pharmacology. 2019 June 15. Handbook of Experimental Pharmacology. |
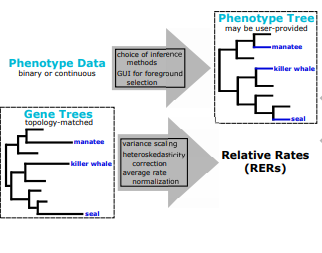 | Kowalczyk A, Meyer WK, Partha R, Mao W, Clark NL, Chikina M. RERconverge: an R package for associating evolutionary rates with convergent traits. 2019 June 13. Bioinformatics. |
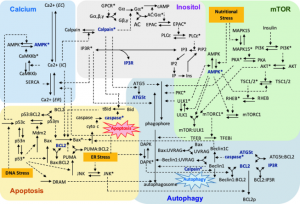 | Liu B, Gyori BM, Thiagarajan PS. Statistical Model Checking-Based Analysis of Biological Networks. 2019 Jun 12. Computational Biology; 30. |
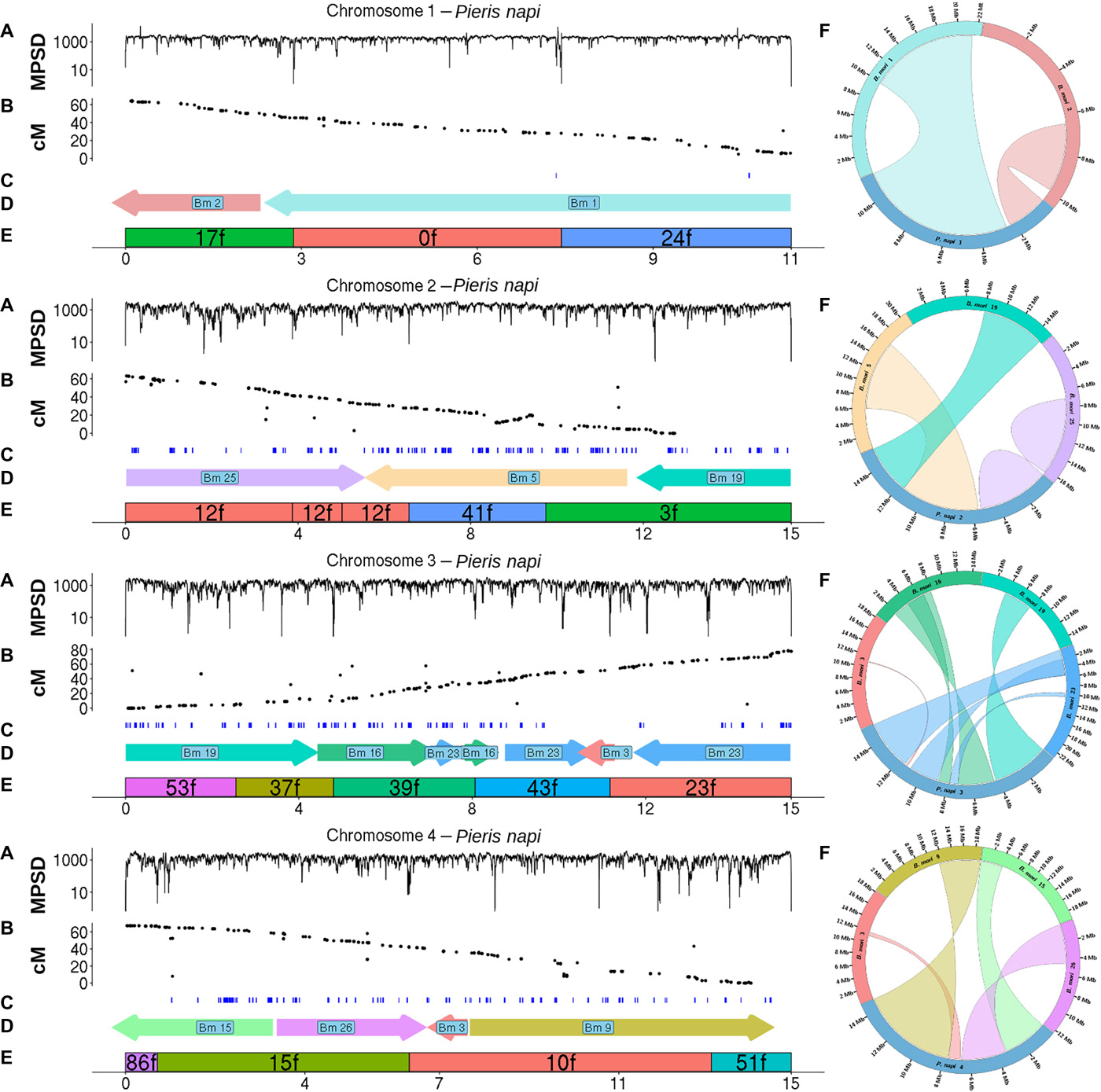 | Hill J, Rastas P, Hornett EA, Neethiraj R, Clark N, Morehouse N, de la Paz Celorio-Mancera M Cols JC, Dircksen H, Meslin C, Keehnen N, Pruisscher P, Sikkink K, Vives M, Vogel H, Wiklund C, Woronik A, Boggs CL, Nylin S, Wheat CW. Unprecedented reorganization of holocentric chromosomes provides insights into the enigma of lepidopteran chromosome evolution. 2019 June 12. Science Advances; 5(6):eaau3648. |
 | Rashid S, Long Z, Sing S, Kohram M, Vashistha H, Navlakha S, Salman H, Oltvai ZN, Bar-Joseph Z. Adjustment in tumbling rates improves bacterial chemotaxis on obstacle-laden terrains. 2019 June 11. Proceedings of the National Academy of Sciences of the United States of America; 116(24): 11770-11775. |
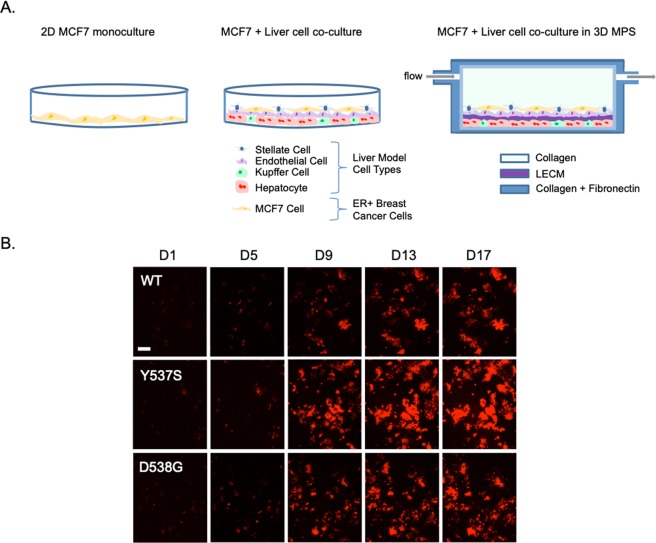 | Miedel MT, Gavlock DC, Jia S, Gough A, Taylor DL, Stern AM. Modeling the Effect of the Metastatic Microenvironment on Phenotypes Conferred by Estrogen Receptor Mutations Using a Human Liver Microphysiological System. 2019 Jun 6. Sci Rep; 9(1):8341. doi:10.1038/s41598-019-44756-5 |
 | Sackton TB,Clark N. Convergent evolution in the genomics era: new insights and directions. 2019 June 3. Philosophical Transactions of the Royal Society B. 374(1777) |
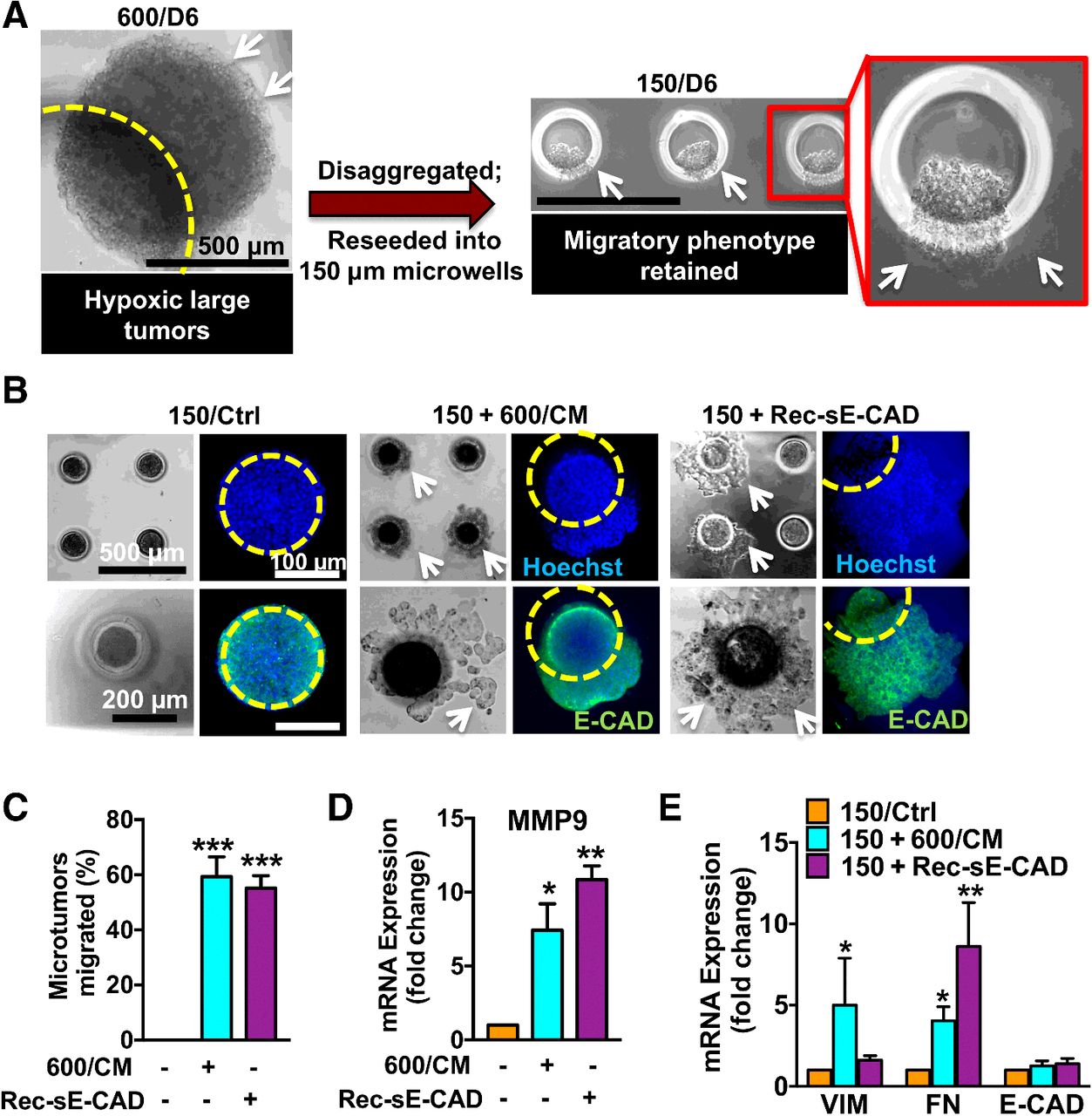 | Singh M, Tian XJ, Donnenberg VS, Watson AM, Zhang J, Stabile LP, Watkins SC, Xing J, Sant S. Targeting the Temporal Dynamics of Hypoxia-Induced Tumor-Secreted Factors Halts Tumor Migration. 2019 June 1. Cancer Research; 79(11):2962-2977. |
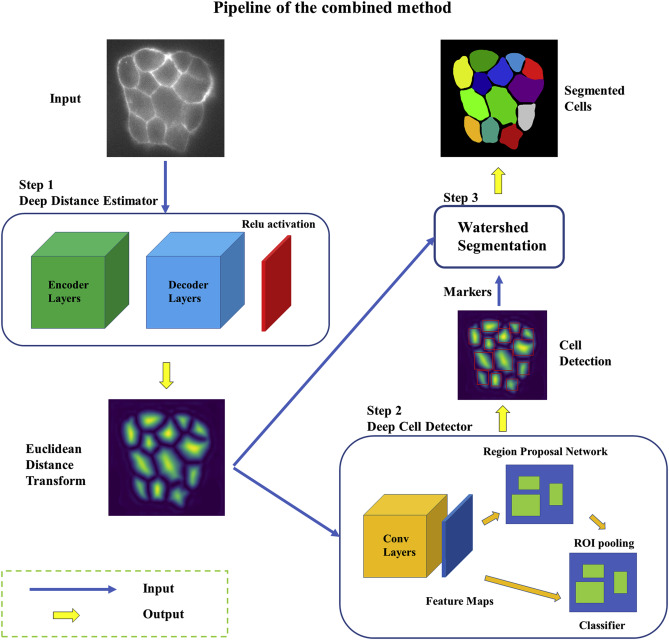 | Wang W, Taft DA, Chen YJ, Zhang J, Wallace CT, Xu M, Watkins SC, Xing J. Learn to segment single cells with deep distance estimator and deep cell detector. 2019 May. Computers in Biology and Medicine; 108:133-141. |
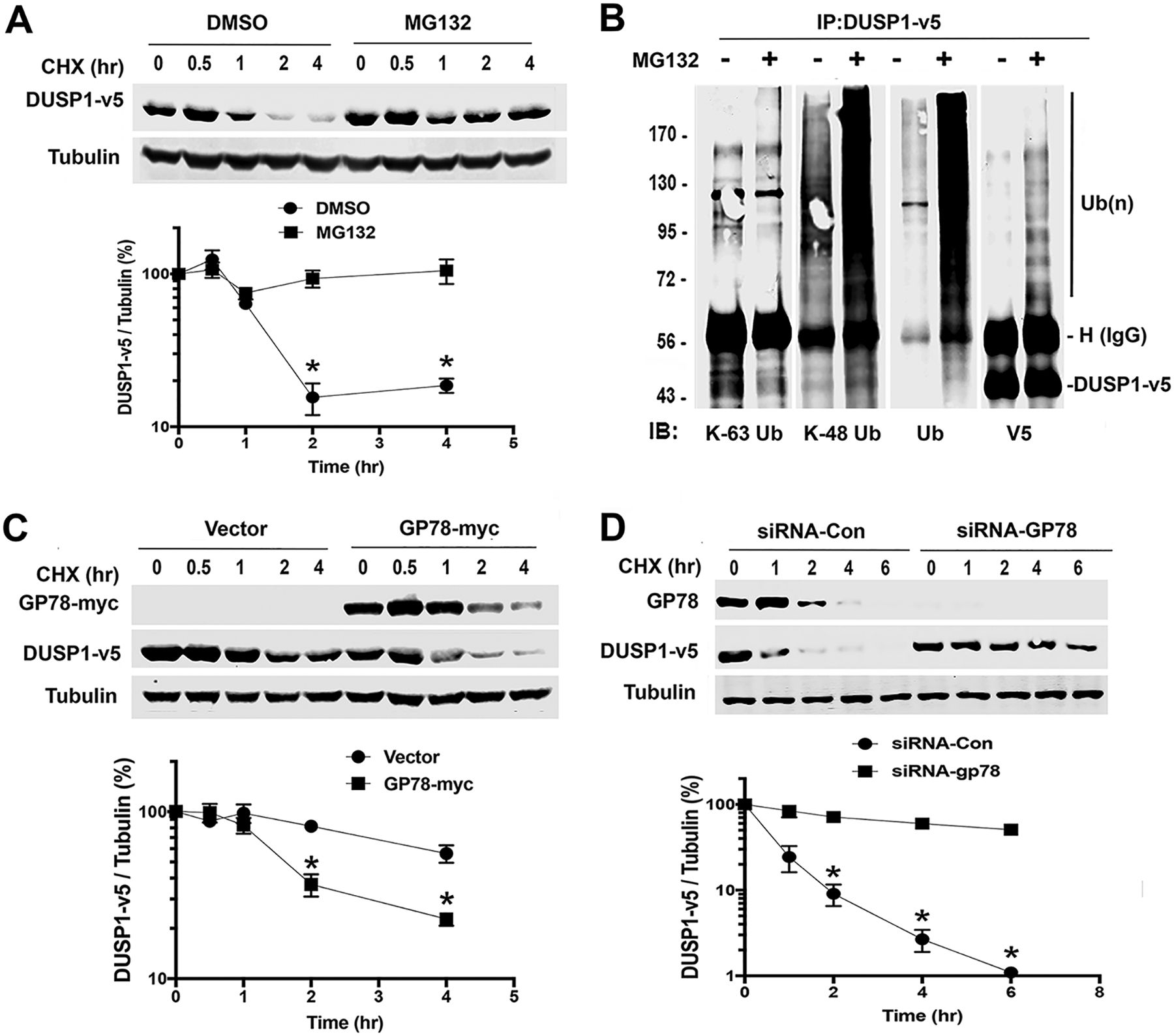 | Kho DH, Uddin MH, Chatterjee M, Vogt A, Raz A, Wu GS. GP78 Cooperates with Dual-Specificity Phosphatase 1 To Stimulate Epidermal Growth Factor Receptor-Mediated Extracellular Signal-Regulated Kinase Signaling. 2019 May 14. Molecular and Cellular Biology; 39(11). |
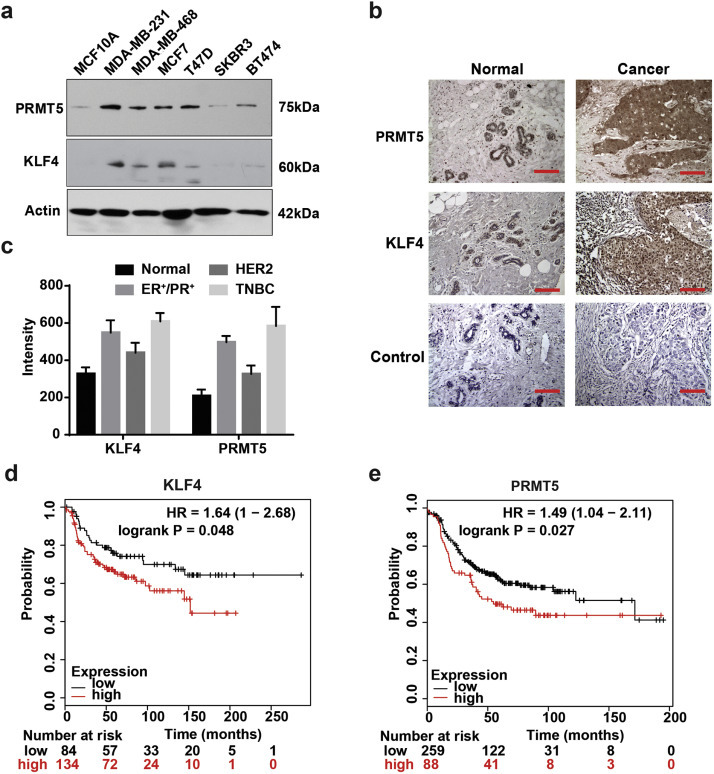 | Zhou Z, Feng Z, Hu D, Yang P, Gur M, Bahar I, Cristofanilli M, Gradishar WJ, Xie X, Wan Y. A novel small-molecule antagonizes PRMT5-mediated KLF4 methylation for targeted therapy. 2019 May 14. EBioMedicine. |
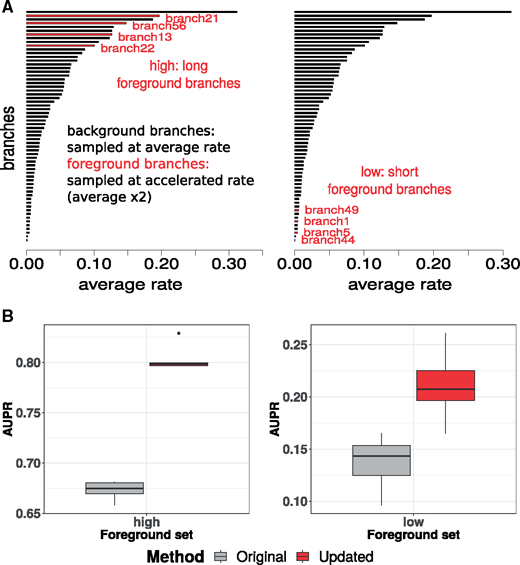 | Partha R, Kowalczyk A, Clark N, Chikina M. Robust Method for Detecting Convergent Shifts in Evolutionary Rates. 2019 May 11. Molecular Biology and Evolution. |
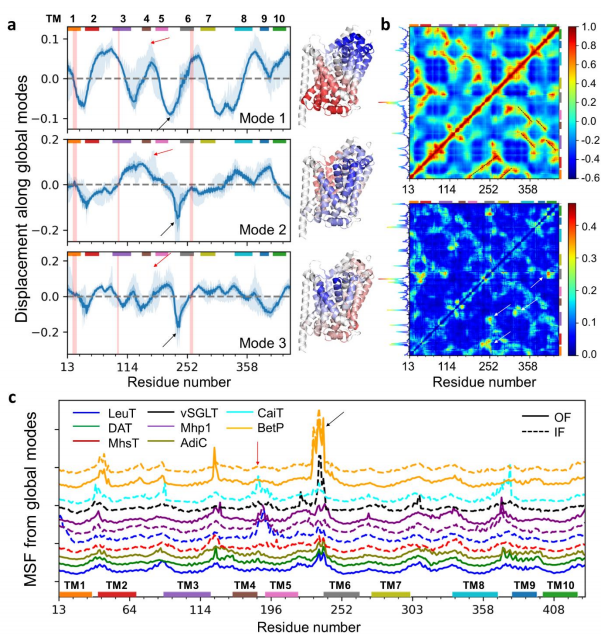 | Zhang S, Li H, Krieger JM, Bahar I. Shared signature dynamics tempered by local fluctuations enables fold adaptability and specificity. 2019 Apr 27. Molecular Biology and Evolution. |
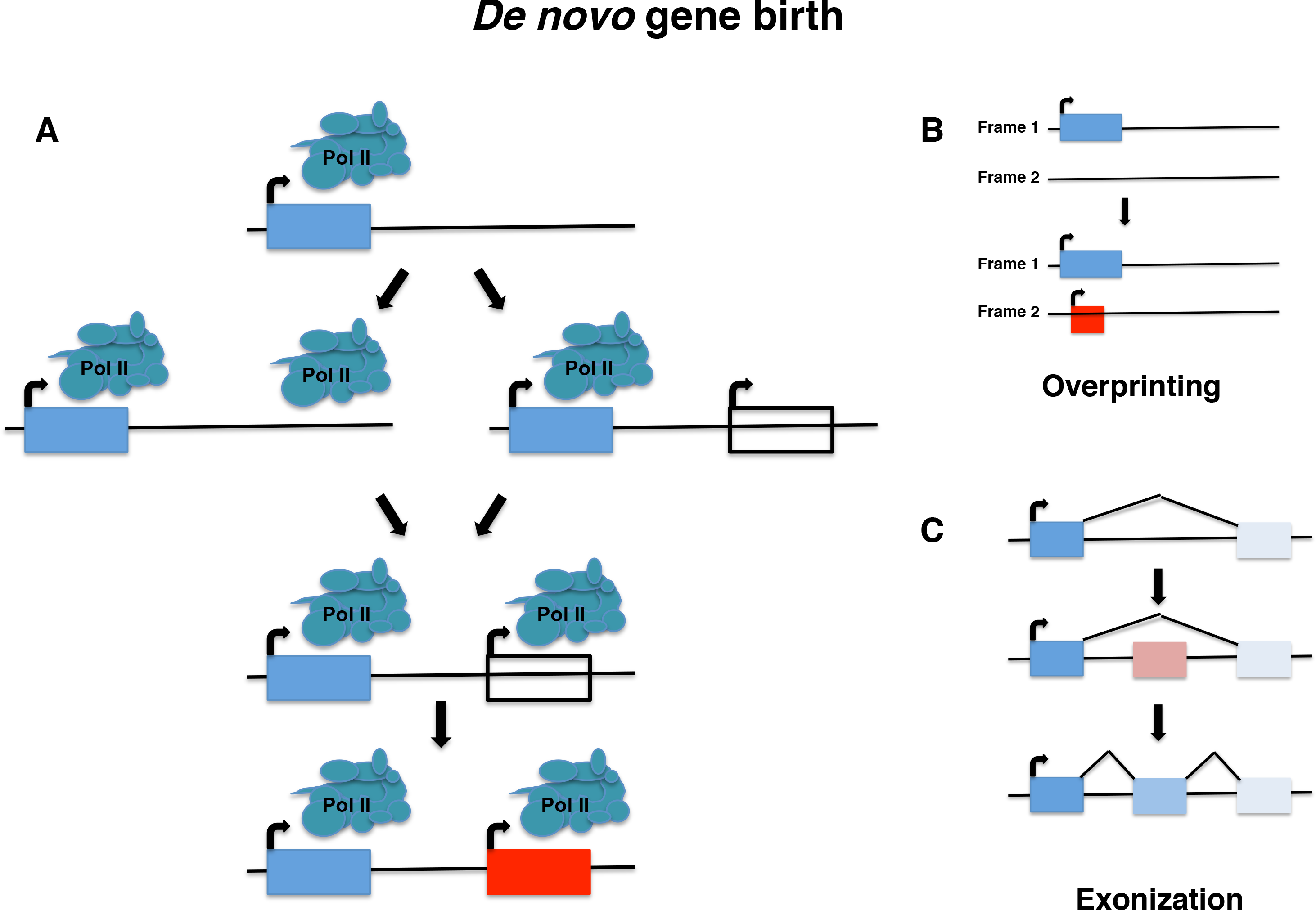 | Van Oss SB, Carvunis AR. De novo gene birth. 2019 May 23. PLoS Genetics; 15(5):e1008160. doi:10.1371/journal.pgen.1008160 |
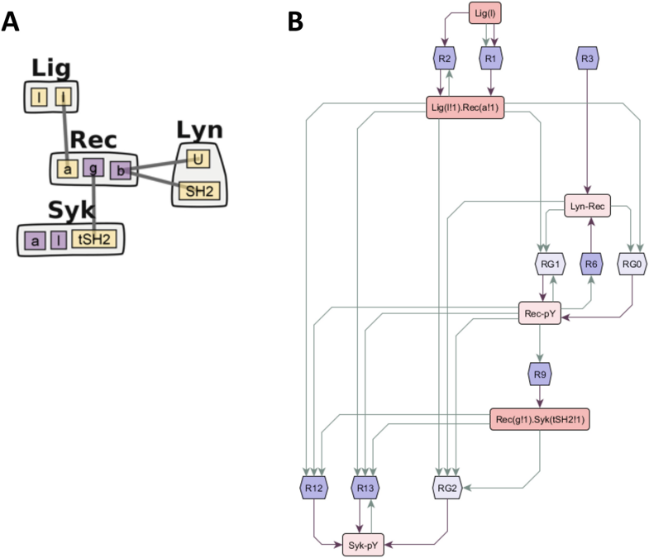 | Tapia J, Saglam AS, Czech J, Kuczewski R, Bartol TM, Sejnowski TJ, Faeder JR. MCell-R: A particle-resolution network-free spatial modeling framework. 2019. Modeling Biomolecular Site Dynamics; 1945: 203-229. |
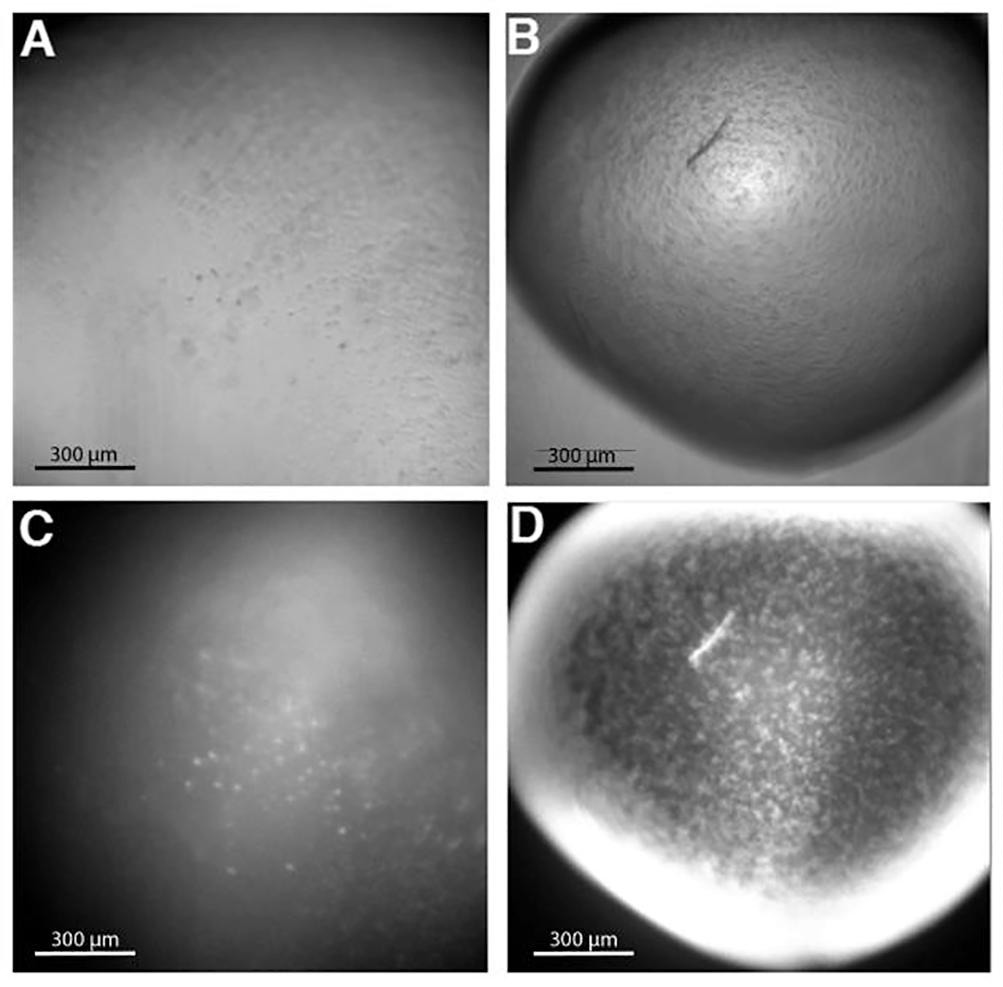 | Guowu L, Weiss SC, Vergara S, Camacho C, Calero G. Transcription with a laser: Radiation-damage free diffraction of RNA Polymerase II crystals. 2019 Apr 25. Methods. |
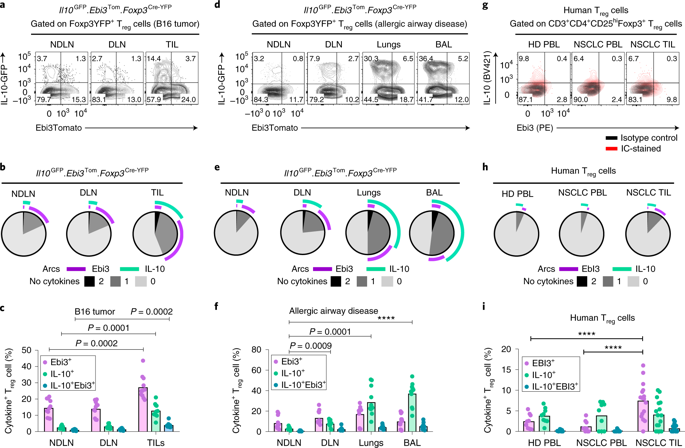 | Sawant DV, Yano H, Chikina M, Zhang Q, Liao M, Liu C, Callahan DJ, Sun Z, Sun T, Tabib T, Pennathur A, Corry DB, Luketich JD, Lafyatis R, Chen W, Poholek AC, Bruno TC, Workman CJ, Vignali DAA. Adaptive plasticity of IL-10+ and IL-35+ Treg cells cooperatively promotes tumor T cell exhaustion. 2019 Apr 01. Nature Immunology. |
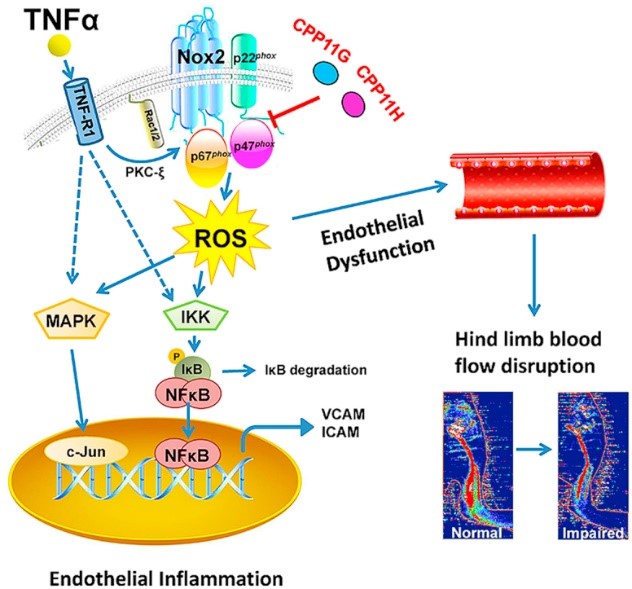 | Li Y, Cifuentes-Pagano E, DeVallance ER, de Jesus DS, Meijles DN, Koes D, Camacho CJ, Ross M, St Croix C, Pagano PJ. NADPH oxidase 2 inhibitors CPP11G and CPP11H attenuate endothelial cell inflammation & vessel dysfunction and restore mouse hind-limb flow. Redox Biol. 2019;22:101143. doi:10.1016/j.redox.2019.101143 |
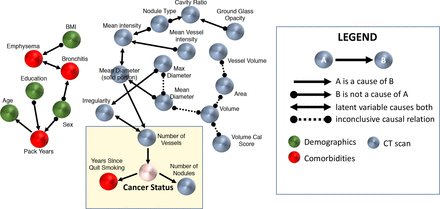 | Raghu VK, Zhao W, Pu J, Leader JK, Wang R, Herman J, Yuan, J, Benos PV, Wilson DO. Feasibility of lung cancer prediction from low-dose CT scan and smoking factor using causal models. Thorax. Published Online First: 12 March 2019. doi: 10.1136/thoraxjnl-2018-212638 |
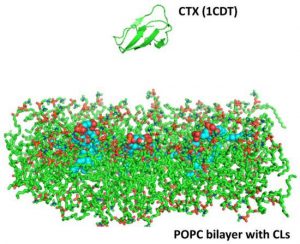 | Zhang B, Li F, Chen Z, Shrivastava IH, Gasanoff ES, Dagda RK. Naja mossambica mossambica Cobra Cardiotoxin Targets Mitochondria to Disrupt Mitochondrial Membrane Structure and Function. 2019 March 8. Toxins; 11(3). |
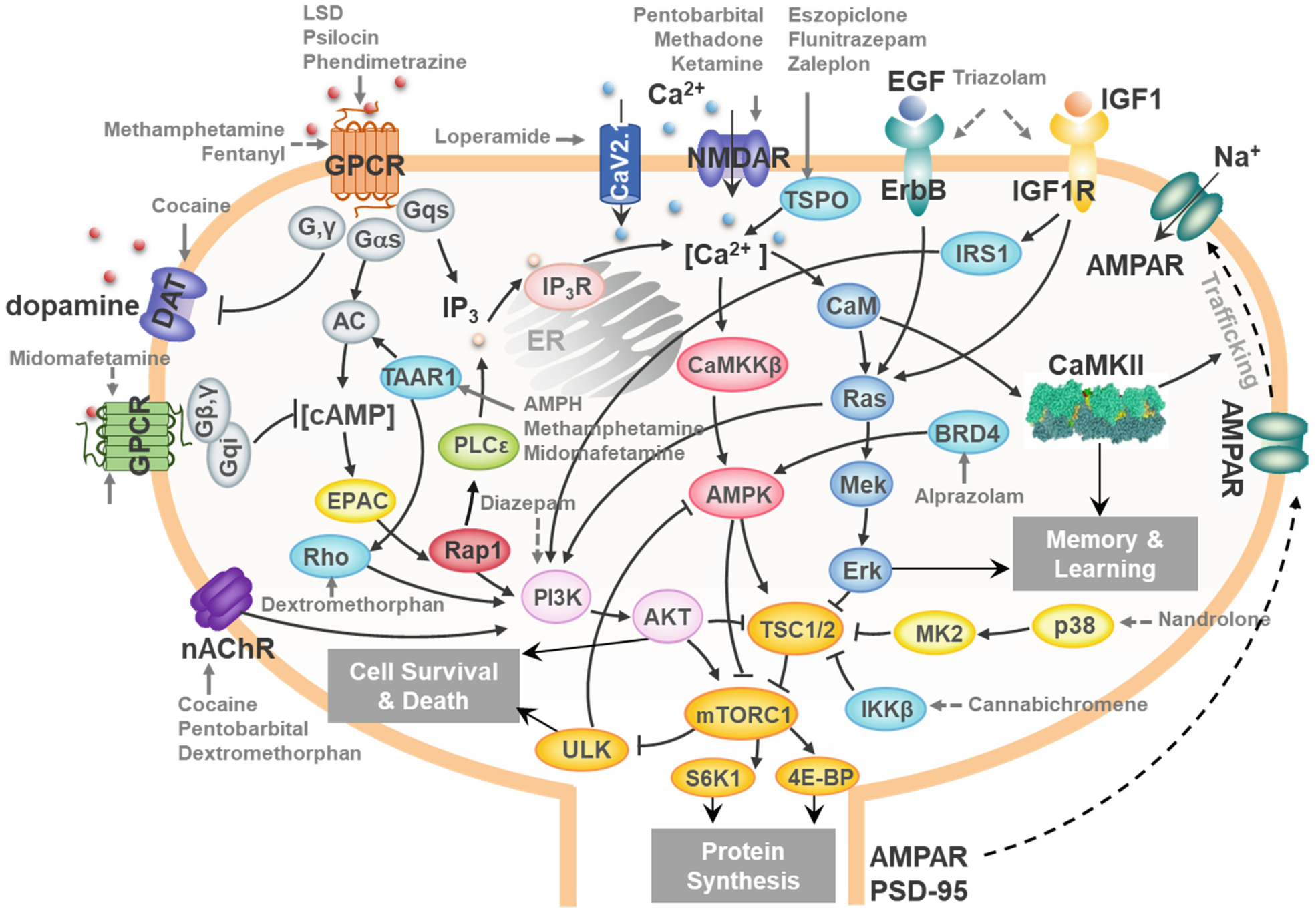
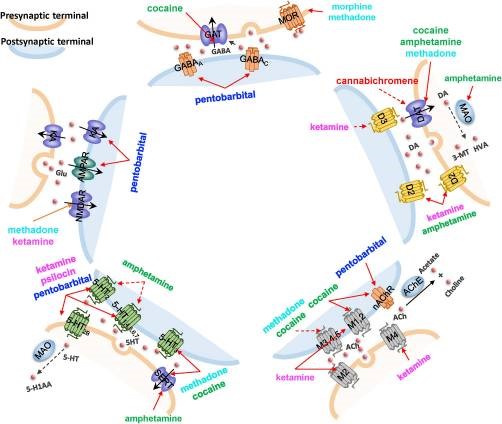 | Pei F, Li H, Liu B, Bahar I. Quantitative Systems Pharmacological Analysis of Drugs of Abuse Reveals the Pleiotropy of Their Targets and the Effector Role of mTORC1. Front Pharmacol. 2019;10:191. Published 2019 March 8. doi:10.3389/fphar.2019.00191 |
 | Xing J, Tian XJ. Investigating epithelial-to-mesenchymal transition with integrated computational and experimental approaches. 2019 March 7. Physical Biology; 16(3):031001. |
 | Raghu VK, Ge X, Balajee A, Shirer DJ, Das I, Benos PV, Chrysanthis PK. (2019) A Pipeline for Integrated Theory and Data-Driven Modeling of Genomic and Clinical Data. BioKDD 2019 |
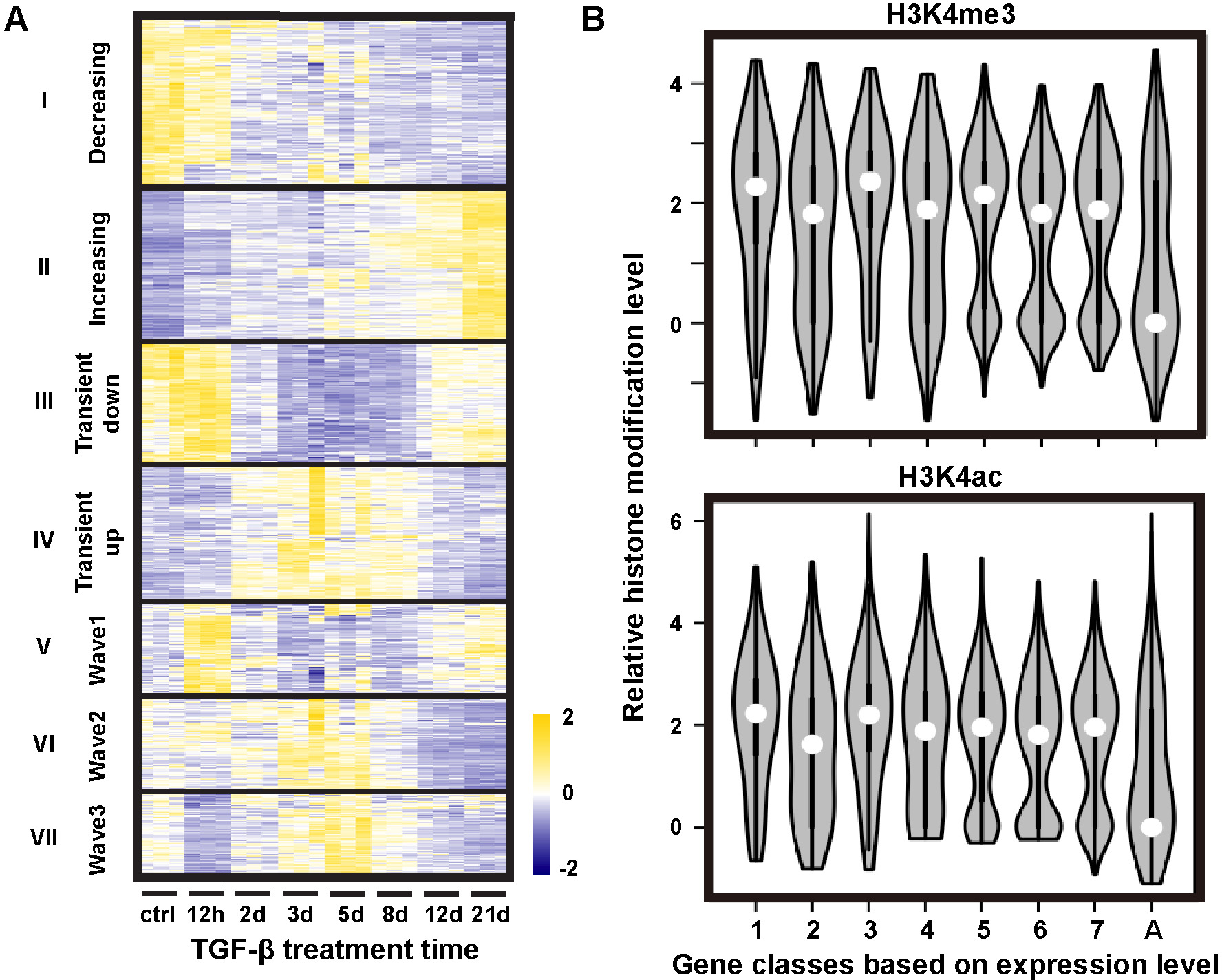 | Zhang J, Chen H, Li R, Taft DA, Yao G, Bai F, Xing J. Spatial clustering and common regulatory elements correlate with coordinated gene expression. 2019 March 1. PLoS Comput Biol. 2019;15(3):e1006786. doi:10.1371/journal.pcbi.1006786 |
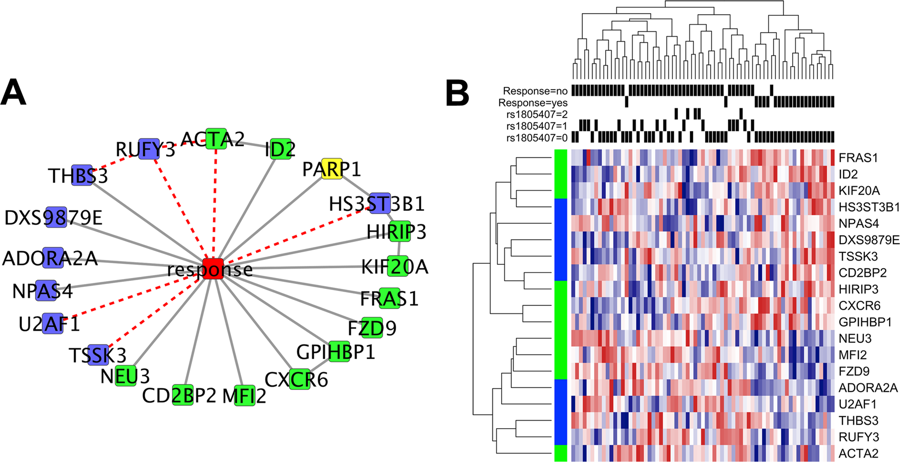 | Abecassis I, Sedgewick AJ, Romkes M, Buch S, Nukui T, Kapetanaki MG, Vogt A, Kirkwood JM, Benos PV. PARP1 rs1805407 Increases Sensitivity to PARP1 Inhibitors in Cancer Cells Suggesting an Improved Therapeutic Strategy. Sci Rep. 2019;9(1):3309. Published 2019 March 1. doi:10.1038/s41598-019-39542-2 |
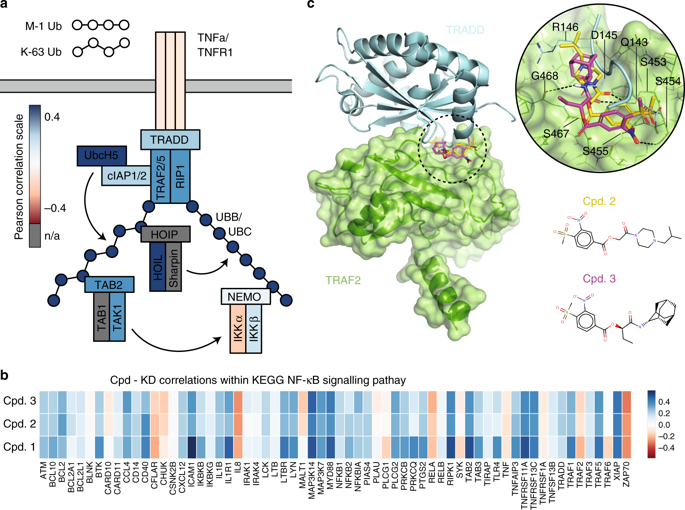 | Pabon NA, Zhang Q, Cruz JA, Schipper DL, Camacho CJ, Lee REC. A network-centric approach to drugging TNF-induced NF-κB signaling. Nat Commun. 2019;10(1):860. Published 2019 Feb 26. doi:10.1038/s41467-019-08802-0 |
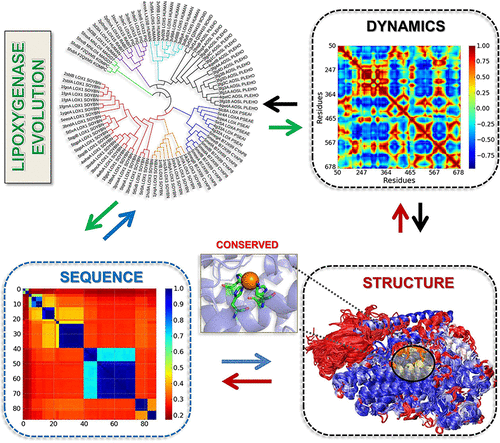 | Mikulska-Rauminska K, Shrivastava I, Krieger, J, Zhang S, Li H, Bayir B, Wenzel SE, VanDemark AP, Kagan VE, Bahar I. Characterization of Differential Dynamics, Specificity, and Allostery of Lipoxygenase Family Members. 2019 Feb 14. Journal of Chemical Information and Modeling. |
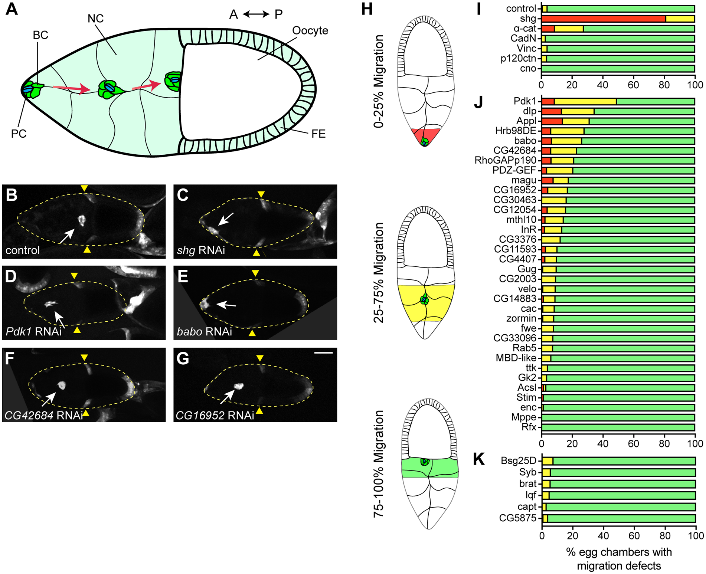 | Raza Q, Choi JY, Li Y, O’Dowd RM, Watkins SC, Chikina M, Hong Y, Clark NL, Kwiatkowski AV. Evolutionary rate covariation analysis of E-cadherin identifies Raskol as a regulator of cell adhesion and actin dynamics in Drosophila. PLoS Genet. 2019;15(2):e1007720. Published 2019 Feb 14. doi:10.1371/journal.pgen.1007720 |
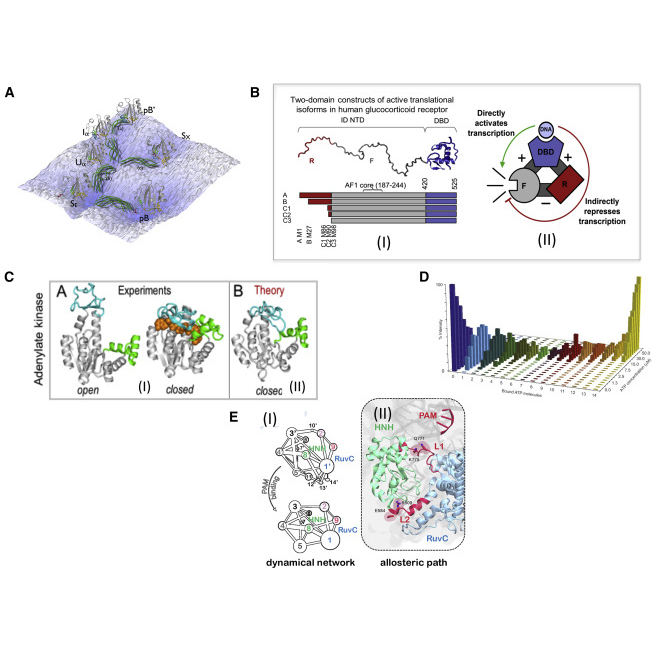 | Wodak SJ, Paci E, Dokholyan NV, Berezovsky IN, Horovitz A, Li J, Hilser VJ, Bahar I, Karanicolas J, Stock G, Hamm P, Stote RH, Eberhardt J, Chebaro Y, Dejaegere A, Cecchini M, Changeux JP, Bolhuis PG, Vreede J, Faccioli P, Orioli S, Ravasio R, Yan L, Brito C, Wyart M, Gkeka P, Rivalta I, Palermo G, McCammon JA, Panecka-Hofman J, Wade RC, Di Pizio A, Niv MY, Nussinov R, Tsai CJ, Jang H, Padhorny D, Kozakov D, McLeish T. (2019) Allostery in Its Many Disguises: From Theory to Applications. Structure. 2019 Apr 2;27(4):566-578. doi: 10.1016/j.str.2019.01.003. Epub 2019 Feb 7. PMID: 30744993 |
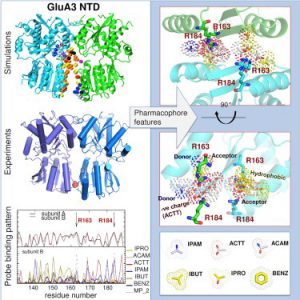 | Lee J Y, Krieger J, Herguedas B, Garcca-Nafrra J, Dutta A, Shaikh SA, Greger IH, Bahar I. (2019) Druggability Simulations and X-ray Crystallography Reveal a Ligand-binding Site in the GluA3 AMPA Receptor N-terminal Domain. Structure. doi:10.2139/ssrn.3188417 |
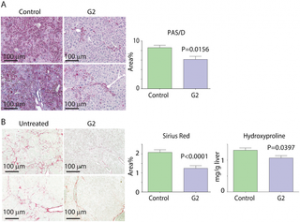 | Wang Y, Cobanoglu MC, Li J, Hidvegi T, Hale P, Ewing M, Chu AS, Gong Z, Muzumdar R, Pak SC, Silverman GA, Bahar I, Perlumtter DH. (2019) An analog of glibenclamide selectively enhances autophagic degradation of misfolded α1-antitrypsin Z. PLoS ONE 14(1): e0209748. https://doi.org/10.1371/journal.pone.0209748 |
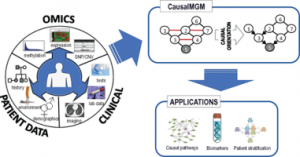 | Sedgewick AJ, Buschur K, Shi I, Ramsey JD, Raghu V, Manatakis DV, Zhang Y, Bon J, Chandra D, Karoleski C, Sciurba FC, Spirtes P, Glymour C, Benos PV. Mixed graphical models for integrative causal analysis with application to chronic lung disease diagnosis and prognosis, Bioinformatics, , bty769, https://doi.org/10.1093/bioinformatics/bty769 |
 | Asan A, Skoko JJ, Woodcock CC, Wingert BM, Woodcock SR, Normolle D, Huang Y, Stark JM, Camacho CJ, Freeman BA, Neumann CA. Electrophilic fatty acids impair RAD51 function and potentiate the effects of DNA-damaging agents on growth of triple-negative breast cells. (2019) Jan 11. Journal of Biological Chemistry, 294(2), 397-404. |
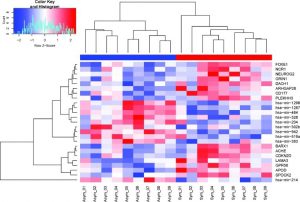 | Ellen M Caparosa, Andrew J Sedgewick, Georgios Zenonos, Yin Zhao, Diane L Carlisle, Lucia Stefaneanu, Brian T Jankowitz, Paul Gardner, Yue-Fang Chang, William R Lariviere, William A LaFramboise, Panayiotis V Benos, Robert M Friedlander; Regional Molecular Signature of the Symptomatic Atherosclerotic Carotid Plaque, Neurosurgery. 2019 Aug 1; 85(2):E284-E293. PMID: 30335165 |
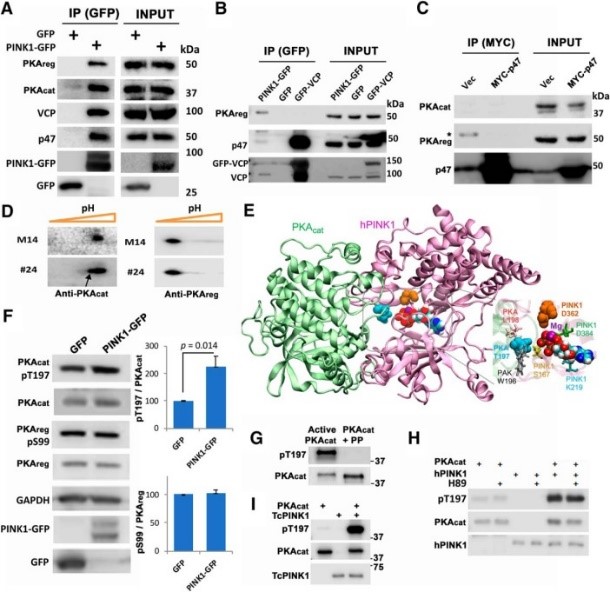 | Wang KZQ, Steer E, Otero PA, Bateman NW, Cheng MH, Scott AL, Wu C, Bahar I, Shih YT, Hsueh YP, Chu CT. PINK1 Interacts with VCP/p97 and Activates PKA to Promote NSFL1C/p47 Phosphorylation and Dendritic Arborization in Neurons. eNeuro. 2018;5(6):ENEURO.0466-18.2018. Published 2018 Jan 10. doi:10.1523/ENEURO.0466-18.2018 |
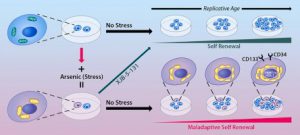 | Cheikhi A, Wallace C, St Croix C, Cohen C, Tang W, Wipf P, Benos PV, Ambrosio F, Barchowsky A. Mitochondria are a substrate of cellular memory. (2019) Free Radical Biology and Medicine, 130, 528-541. https://doi.org/10.1016/j.freeradbiomed.2018.11.028 |
 | Sunseri J, King JE, Francoeur PG, Koes DR. Convolutional neural network scoring and minimization in the D3R 2017 community challenge. 2019. Journal of Computer-Aided Molecular Design; 33(1):19-34. |
Other Years
‣ 2021 Publications
‣ 2020 Publications
‣ 2019 Publications
‣ 2018 Publications
‣ 2017 Publications
‣ 2016 Publications
‣ 2015 Publications
‣ 2014 Publications
‣ 2013 Publications
‣ 2012 Publications
‣ 2011 Publications
‣ 2010 Publications
‣ 2009 Publications
‣ 2008 Publications
‣ 2007 Publications
‣ 2006 Publications
‣ 2005 Publications
